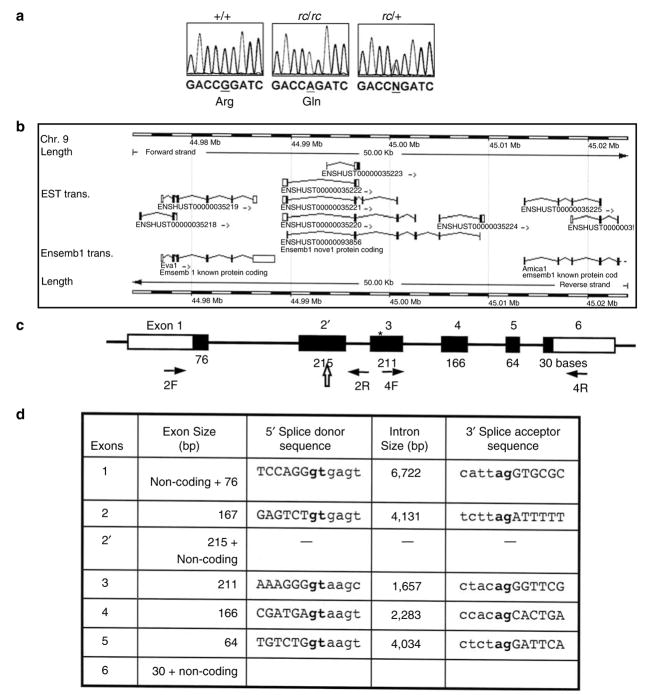
Figure 4. Gene structure and mutation analysis of a novel gene in which a mutation was identified in the rc/rc DNA.
(a) Sequence analysis identified a point mutation in a predicted open reading frame in the rc/rc genomic DNA, which was confirmed in rc/+ genomic DNA and rc/rc cDNA. We have named this gene “Mpzl3” based on the similarity of the predicted domain structure of its encoded protein to MPZ and MPZL2 (also called EVA1). (b) The Mpzl3 gene (ENSMUSG00000070305), predicted on the basis of multiple Ensembl transcripts based on ESTs, in its genomic context. Eva1 (Mpzl2) gene is upstream, and Amica1 gene is downstream. (c) Our prediction of the murine Mpzl3 gene that is homologous to the human gene ENSG00000160588 and rat gene ENSRNOESTG00000015598. Exon and EST information was based on the Ensembl Genome Database (www.ensembl.org) and our prediction by sequence comparison between species. Numbers underneath each exon indicate the length of the coding sequence in that exon (filled portion). Exon 2 has 167 bp of coding sequence, and exon 2′ has 215 bp of coding sequence (48 bp longer than exon 2) as well as a 3′ UTR. Block arrow points to the splice site in exon 2′ used to generate the six-exon transcript. The arrows denote primers 2F, 2R, 4F, and 4R used for RT-PCR analysis in Figure 6. *The position of the mutation detected in rc/rc DNA. (d) Exon–intron information of the predicted murine Mpzl3 gene.