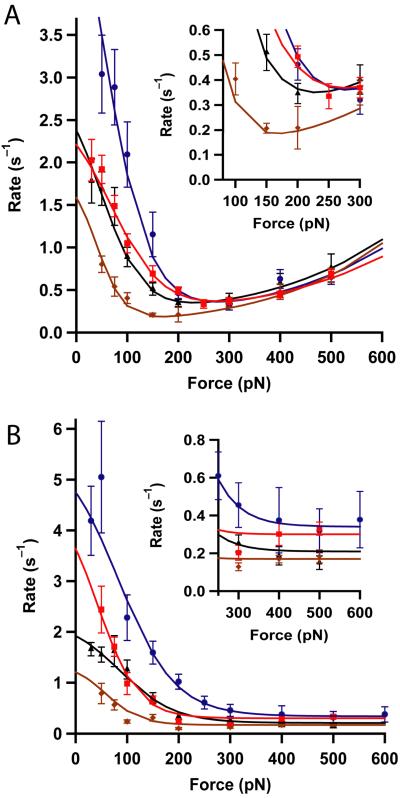
Figure 3. Force-dependency of the rate of disulfide reduction by Trx enzymes from different species.
(A) Bacterial-origin Trxs: human mitochondrial Trx2 (blue), E. coli Trx1 (red), pea chloroplastic Trxm (black), E. coli Trx2 (brown). While the Michaelis-Menten (low force) mechanism differs in magnitude among the Trxs, the simple SN2 like reaction observed at higher forces is very similar in all of them. The inset shows a magnified view of the traces in the region where they reach a minimum. (B) Eukaryotic-origin Trxs: human Trx1 (blue, from ref 11), Plasmodium Trx (red), poplar Trxh1 (black), and poplar Trxh3 (brown). The most noticeable feature is the absence of the SN2 like reaction at high forces in all eukaryotic Trxs. The inset shows an expansion of the minimum rate of reduction attained at high forces. In all experiments the concentration of Trx was 10 μM. The smooth lines are fits of the kinetic model described in the supplementary information. The kinetics parameters obtained are summarized in Table 1. The error bars are given by the standard error of the mean obtained with the bootstrap method.