Abstract
Reversible and easy to use, temperature-sensitive (TS) mutations are powerful tools for studying gene function. However, TS alleles are rare and difficult to generate and identify, and this has limited their use in most multicellular organisms. We have generated and characterized 41 intein switches, temperature-sensitive Sce VMA mutations that splice only at the permissive temperatures to generate intact host proteins. At nonpermissive temperatures, they fail to splice, resulting in a loss of function of the proteins in which they reside. By inserting an intein switch into a protein of interest, one can turn on and off the activities of the engineered protein with a simple temperature shift. The 41 TS inteins function in five different temperature ranges, with permissive temperatures ranging from 18° to 30°. This collection makes it possible to choose a TS-intein switch according to the optimal growth temperature of an organism or to suit a special experimental design.
LOSS-OF-FUNCTION phenotypes provide critical insights into gene functions. Conventional gene-targeting techniques generate loss-of-function mutations by permanently deleting the specific gene of interest or by rendering it nonfunctional. However, this strategy falls short for two groups of genes: essential genes and pleiotropic genes. Essential genes are required for viability and account for about a quarter of the genes in various organisms, including mice, flies, worms, and yeast (Miklos and Rubin 1996). Pleiotropic genes, to which the vast majority of genes belong, function at multiple times and/or multiple places during the life cycle of an organism.
In contrast to conventional gene knockouts, heat-sensitive mutations, traditionally known as temperature-sensitive (TS) mutations, are powerful tools for studying the functions of all genes. Much of our understanding of the fundamental aspects of cell division has relied on the analysis of TS mutations in yeast (Pringle 1975). In Drosophila melanogaster, the use of a TS allele of Notch allowed its various spatial and temporal functions to be defined (Shellenbarger and Mohler 1978; Presente et al. 2001; Costa et al. 2005). TS mutations are functional at low (permissive) temperatures, yet nonfunctional at high (nonpermissive) temperatures, and thus a rise in temperature quickly ablates protein function. This approach is intrinsically specific; only the protein that is engineered to be conditional is targeted under nonpermissive temperatures. In addition, TS mutations are versatile and convenient to use. However, the scarcity of TS alleles and the difficulty of generating and identifying them have limited their use (Suzuki et al. 1971; Harris and Pringle 1991), especially in multicellular organisms.
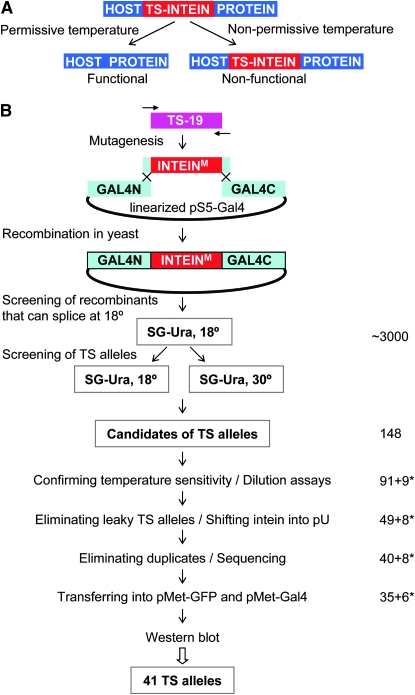
We previously described a method that utilizes a conditionally splicing intein, an intein switch, to generate TS mutations (Zeidler et al. 2004). An intein is a self-excising stretch of amino acids, which is removed during protein maturation (Hirata et al. 1990; Kane et al. 1990). An intein switch splices itself only at permissive temperatures to generate an intact host protein (Figure 1A). At nonpermissive temperatures, it fails to splice and remains within the host protein, leading to the inactivation of that protein. Intein splicing, also known as protein splicing, is a post-translational process in which the intein is precisely excised from a nascent protein precursor, and the two flanking sequences (N and C exteins) are ligated together through a normal peptide bond (Evans and Xu 2002; Anraku et al. 2005; Perler 2005). Thus, no intein footprint is left behind after protein splicing; that is, whatever allele a host protein possesses, be it wild type, dominant negative or constitutively active, the same allele will be generated after splicing. Furthermore, protein splicing is a self-catalytic reaction that does not require any exogenous proteins, cofactors, or energy sources. This makes protein splicing applicable to any organism, and the naturally occurring inteins found in unicellular organisms can be used in multicellular organisms.
Figure 1.—
Generation and characterization of TS inteins. (A) Principle of TS-intein function. (B) Flowchart of steps used to generate the second-generation TS inteins and to characterize the TS-intein mutations from both the first and second generation. See text for details. Numbers on the right represent the number of TS alleles retained at each step. An asterisk indicates the number of first-generation TS-intein alleles retained at each step.
The first generation of TS inteins was generated by random mutagenesis of wild-type Saccharomyces cerevisiae vacuolar membrane ATPase intein (Sce VMA) (Zeidler et al. 2004). These TS inteins have been used in bacteria, yeast, flies, and zebrafish (Zeidler et al. 2004; Liang et al. 2007; S. Holley, personal communication) to conditionally control protein activities, demonstrating the generality of this approach. However, these TS inteins have not been fully characterized. More importantly, 30°, the nonpermissive temperature of the first-generation TS inteins is too high for long-term incubation of certain organisms. For example, a temperature of 30° is stressful for D. melanogaster, and long-term incubation at 30° greatly reduces its fertility and life span. In addition, different organisms have different optimal growth temperatures. These concerns prompted us to generate TS-intein alleles that function at a variety of temperature ranges.
We have generated a large collection of new TS-intein alleles by mutagenizing one of the first-generation TS-intein alleles, F19 (originally known as TS19). To facilitate the application of the TS inteins, we characterized both the first- and the second-generation TS inteins. A total of 41 (35 newly generated) TS-intein alleles were retained after characterization. They were classified into five groups according to their permissive temperatures—group I: 18°; group II: 22°; group III: 25°; group IV: 28°; and group V: 30°. All 41 alleles spliced in a temperature-dependent manner in the two host proteins tested. The alleles with higher permissive temperatures were able to splice at higher temperatures, and the vast majority behaved in the same way in both host proteins. Time-course studies of the splicing of group II to group V TS inteins showed that significant splicing occurred within 1–2 hr at 18°. This expanded and characterized collection of TS inteins makes it possible to choose a TS-intein switch on the basis of the optimal growth temperature of an organism or tailored to a specific experimental design. These TS-intein alleles can be used to confer temperature sensitivity on proteins in a variety of organisms, thus facilitating the functional annotation of genomes.
MATERIALS AND METHODS
Yeast strains and culturing:
Yeast strains used in this experiment were S. cerevisiae FY760 (MATa ura3-52 trp1Δ63 leu2Δ1 gal4Δ∷LEU2 his3Δ200 lys2-128Δ) (Zeidler et al. 2004) and S. cerevisiae FY761 (MATa ura3-52 trp1Δ63 leu2Δ1 gal4Δ∷LEU2 his3Δ200 lys2-128Δ Sce VMAΔ), which was derived from FY760 by deletion of Sce VMA and mutation of the Sce VMA homing site (changed from catcatctatgtcgggtgcggagaaagaggtaatgaatggc to catcatttacgtcggttgtggtgagaggggaaacgagatggc; new bases are in boldface).
Yeast transformation was performed as described in Gietz and Woods (2002). Yeast dilution assays were eightfold dilution series on uracil drop-out synthetic media with 2% dextrose (SD-Ura) or with 2% galactose (SG-Ura). Plates were incubated at the indicated temperatures and examined after 3–6 days.
Plasmids and mutagenesis:
Plasmids pS5-Gal4 and pU-Gal4 have been reported previously (Zeidler et al. 2004). Plasmid p972 (Pinson et al. 1998) was a gift from B. Daignan-Fornier (Institut de Biochimie et Genetique Cellulaires), and pFA6a-GFP(S65T)-kanMX6 (Wach et al. 1997) was a gift from F. Winston (Harvard Medical School).
Intein mutagenesis was done by low-fidelity PCR as described in Cadwell and Joyce (1992) using ExTaq DNA polymerase (TaKaRa), template pU-Gal4-inteinF19, and the primers tgcgatatttgccgacttaaaaagcttaaatgctttgcca and acttggcgcacttcggtttttctttggagcaattatggac. The PCR products were incorporated into GAL4 (between bases 60 and 61) by gap repair in yeast as described in Raymond et al. (1999) and Zeidler et al. (2004) and screened on SG-Ura plates at 18°. About 3000 clones were picked and resuspended in 400 μl water, and 2 μl were dotted onto each of two sets of SG-Ura plates. One set was incubated at 18° and the other at 30°. The colonies containing a candidate of the TS-intein allele grew at 18° but not at 30°. Plasmids were recovered from yeast cultures as described in Hoffman and Winston (1987) and transformed into Escherichia coli Top10 (Invitrogen) by electroporation.
To transfer TS inteins from pS5 to pU, TS intein and GAL4 were released by XhoI and BamHI from the pS-Gal4 intein and ligated with pU-Gal4 digested with the same enzymes. To transfer TS inteins from the pS5-Gal4 intein to p416-Met25 [originally described in Mumberg et al. (1994); the one we used was derived from p972 (Pinson et al. 1998)], Gal4 inteins were released from the pS5-Gal4 intein by KpnI digestion, T4 DNA polymerase blunting, and BamHI digestion and were then ligated with p416-Met25 that had been digested with EcoRI, blunted with T4 DNA polymerase, and digested with BamHI. p416-Met25-eGFP inteins, into which intein was incorporated into eGFP after amino acid 108, were constructed by gap repair in yeast as described in the supporting information, Figure S1. K108A T109C was introduced into eGFP as described in Buskirk et al. (2004). p416-Met25 BasGFP inteins were constructed via a three-way ligation by combining a 2-kb BamHI–XbaI PCR fragment containing eGFP inteins (wild type, dead, or TS4), with a 2.5-kb XbaI–SalI fragment containing the BAS1 coding region excised from p972 (Pinson et al. 1998) and the 5.4-kb BamHI–SalI fragment containing the plasmid backbone (met25 promoter, ampicillin resistance gene, and URA3 gene) from p972. The oligonucleotides used to generate the GFP intein 2-kb BamHI–XbaI PCR fragment were attggatccgcggccgcgaattcatgtctttaattaacagtaaaggagaa and tgtgtaccgtacctacttgatatgtttagatcttga.
Protein expression and extraction:
Yeast carrying a p416-Met25-Gal4 intein or p416-Met25-eGFP intein was cultured in uracil drop-out synthetic media with 2% dextrose and 400 μm methionine (SD-Ura+Met) until 0.6 OD, spun down, washed twice with doubly distilled water, and then resuspended in SD-Ura-Met media (0.7 OD) to induce transcription. After 30 min incubation at 30°, sterile 100× methione was added to a final concentration of 400 μm to stop transcription. The yeast cultures were then separated into 1-ml aliquots in 1.5-ml Eppendorf tubes. These tubes were incubated at different temperatures to allow for intein splicing.
Yeast proteins were extracted with glass beads and Thorner buffer [8 m urea, 5% SDS, 50 mm Tris–HCl (pH 6.8), 5% β-mercaptoethanol] (Sambrook et al. 1989) containing protease inhibitor cocktail (Roche, complete protease inhibitor cocktail tablet). Yeast cells were first spun down at 7500 rpm (Eppendorf, centrifuge 5415D) for 1 min, and the pellets were then resuspended in 60 μl of Thorner buffer preheated to 70°. Immediately after the yeast cells were resuspended, they were transferred into another tube that was preloaded with 30 mg of glass beads and that was also preheated to 70°. This process was completed as quickly as possible to stop protein splicing as well as to inactivate endogenous protease activity. After 2 min at 70°, the samples were vortexed at high speed for 4 min, and the tubes were transferred back to the 70° for 5 more minutes to denature the proteins. After that, the debris was spun down for 2 min at 13,200 rpm (Eppendorf, centrifuge 5415D), and the supernatant was transferred to a fresh tube. The proteins were then stored at −80° until they were analyzed by Western blotting.
Western blotting:
Western blot analyses were carried out using standard methods (Sambrook et al. 1989). Antibody dilutions were 1:4000 in PBS with 0.1%Tween for anti-GFP (Sigma G1544); 1:5000 for both anti-Gal4p (Sigma G9293) and anti-rabbit IgG HRP-conjugated antibody (Invitrogen G21234). The membranes were developed with enhanced chemiluminescence reagent (Pierce #32106) and imaged with the Fujifilm LAS-3000 imaging system.
Time course:
To study the kinetics of intein splicing, 1 ml of yeast cells expressing a BasGFP intein (from the p416-Met25-BasGFP intein, where the intein is located at the same position in eGFP as in eGFP alone) or an eGFP intein (from the p416-Met25-eGFP intein) was harvested at different time points, lysed, and analyzed by Western blotting using anti-GFP antibody. For the BasGFP intein, transcription induction was performed at the same temperatures as the splicing temperatures. For the GFP intein, all transcription induction was performed at 30°. Western blots were quantified with ImageJ (http://rsbweb.nih.gov/ij/). Microsoft Excel was used to perform linear regression and to calculate splicing halftime, assuming that the splicing is a first-order reaction.
RESULTS
Generation of TS-intein alleles that function within different temperature ranges:
New TS-intein alleles were generated by mutagenizing F19 and screening for the abilities of their GAL4 fusions to rescue the growth of GAL4-deleted yeast strain FY760 on galactose media at various temperatures (outlined in Figure 1B). Briefly:
Step 1: To generate and screen for intein mutations that were able to splice at 18°. F19 was mutagenized by low-fidelity PCR. In each PCR primer, 30 bp of GAL4 sequence that flanks the intein insertion site was included so that the mutated intein could be cloned into GAL4 by homologous recombination in yeast (Raymond et al. 1999; Zeidler et al. 2004). The PCR products were cotransformed with linearized pS5-Gal4 into FY760. pS5 is a low-copy-number (one or two copies per yeast cell) yeast–bacteria shuttle vector, which ensures that only one intein allele exists in each yeast clone. The transformants were plated on uracil drop-out synthetic media with 2% galactose (SG-Ura) and cultured at 18° to select for successful recombination and functional Gal4p. Since the only carbon source for yeast growth was galactose, whose metabolism requires functional Gal4p, only yeast containing pS5-Gal4 with intein mutations that spliced at 18° were able to grow.
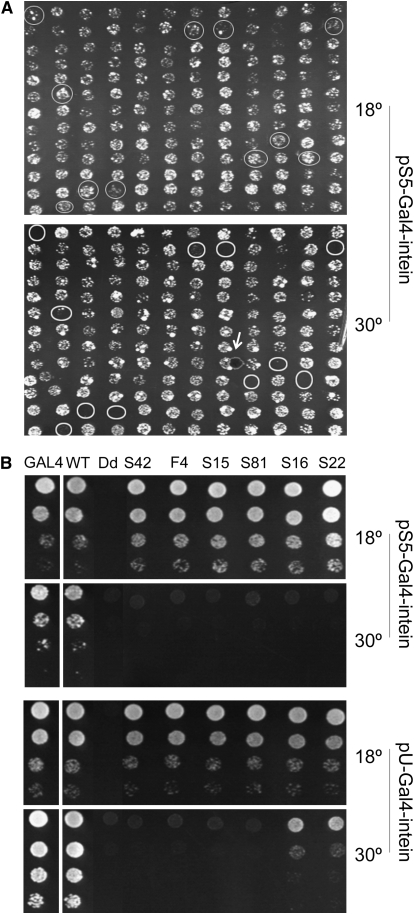
Step 2: To screen for mutations that made yeast growth temperature dependent. About 3000 colonies were picked and plated, in duplicates, onto two sets of SG-Ura plates. One set was incubated at 18°, and the other at 30° (Figure 2A and data not shown). We then screened for colonies that would grow at 18° but not at 30° and found 148 such colonies.
Step 3: To determine whether the candidates were truly TS. Yeast dilution assays were performed, in which serial 1:8 dilutions of yeast were arrayed onto plates, with the highest concentration at 0.1 OD (optical density at 600 nm), and the growth of yeast was monitored. Fifty-seven were discarded at this step either because they grew at 30°, albeit more weakly than at 18°, or because they grew only weakly at 18°.
Step 4: To eliminate TS alleles with leaky splicing at 30°. The 91 confirmed TS-intein alleles were transferred from the pS5 vector into the high-copy-number (∼20 plasmids/yeast cell on average) vector pU and analyzed with the yeast dilution assay. Since more Gal4-intein transcript and protein were produced in yeast carrying multiple copies of the pU-Gal4 intein than in yeast with one or two copies of the pS5-Gal4 intein, the leakiness of TS-intein splicing at nonpermissive temperatures was amplified when expressed in the pU vector (Figure 2B and data not shown). For example, all the TS-intein alleles in Figure 2B behaved similarly in pS5, allowing the yeast transformed with them to grow well at 18° but not at 30°. However, in pU, alleles S16 and S22 enabled their host yeast to grow at 30°, and thus were discarded. Forty-two alleles were eliminated at this step because of leaky splicing at 30°.
Step 5: To eliminate duplicate TS alleles. The remaining 49 alleles were sequenced, and 40 unique alleles were identified (Figure S2). No obvious pattern of mutations emerged from the analysis of the primary peptide sequences of these TS-intein alleles.
Figure 2.—
Identification of TS inteins that splice only in a limited temperature range. (A) Screening for the second-generation TS inteins. About 3000 clones were screened (156 are shown). Colonies that grew well at 18° (top) but not at 30° (bottom) are circled. (The small circle indicated by an arrow in the bottom panel is a bubble.) (B) Eliminating leaky TS-intein alleles. Images show the growth profiles of yeast rescued by GAL4 containing selected intein alleles expressed from the low-copy-number plasmid pS5 (top) and the high-copy-number plasmid pU (bottom). Each row represents an eightfold dilution of the previous concentration. S16 and S22 behaved like other TS-intein alleles at 18°, but supported much better yeast growth at 30° than other mutations did. Note that Gal4-inteinWT (second column) allowed yeast to grow equally well as Gal4p without the intein inserted (first column), while inteinDd (third column) made Gal4p nonfunctional, regardless of the temperature.
The 9 first-generation TS-intein alleles were also included in the pU-yeast dilution assay at step 4 and were further characterized together with the newly generated alleles (Figure 1B). The first- and the second-generation TS-intein alleles were distinguished by their prefixes: “F” for first and “S” for second (Figure 2B and Figure 3). F17, similar to S16, allowed its host yeast to grow at 30°, although not as well as yeast with wild-type intein, and thus was not further characterized. On the basis of this dilution assay, 48 alleles were retained, of which 7 were not included in further analyses due to cloning problems. This resulted in a total of 41 TS-intein alleles, 35 of which were newly generated.
Figure 3.—
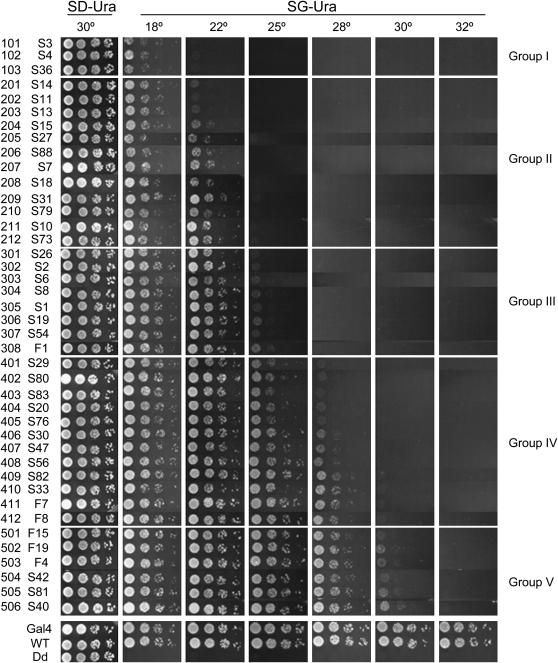
Growth profiles of yeast FY761 transformed with different pU-Gal4 intein alleles. From top to bottom, intein alleles are listed in order of increasing permissive temperatures, except for controls at the very bottom. Note that the yeast at the bottom grew better than those at the top. Although the transition from one allele to the next is gradual, for the convenience of description, we grouped the TS-intein alleles into five groups. Each row in a panel represents a 1:8 dilution series for the specified intein allele. SD-Ura: synthetic media lacking uracil, dextrose as sugar source; SG-Ura: synthetic media lacking uracil, galactose as sugar source; WT: GAL4 with wild-type intein inserted; Dd: GAL4 with dead intein inserted; F1 to F19 are first-generation TS-inteins; S1 to S88 are second-generation TS-inteins; three-digit numbers in all figures: ID numbers for the TS inteins.
Intein alleles function in five different temperature ranges:
To distinguish the optimal functioning temperatures for the various TS inteins, we tested their ability to support the growth of yeast strain FY761 transformed with pU-Gal4 TS inteins at more refined temperatures. FY761 was derived from FY760 by deleting its genomic Sce VMA and by mutating the Sce VMA homing site (G. Tan and C. Tan, unpublished results). FY761 was used to avoid the occasional interference of the endogenous genomic Sce VMA.
As expected, yeast with any of the 41 pU-Gal4 TS inteins grew equally well on glucose media (Figure 3, the first column). In contrast, on galactose media, yeast grew at the temperatures determined by the intein alleles that they carried (from the second column onward). Yeast with wild-type intein (inteinWT) grew at all temperatures; those with dead intein (inteinDd), which is unable to splice from its host protein, did not grow at any temperature; and the TS inteins conferred temperature-dependent growth. The 41 TS-intein alleles differed in their ability to support the galactose-dependent growth of FY761 at different temperatures and were classified into five groups. Yeast with group I TS inteins grew well at 18° but not at >22°; those with group II grew at ≤22°, but not >25°; those with group III grew at ≤25°, but not >28°; those with group IV grew at ≤28°, but not >30°; and those with group V grew at ≤30°, but not >32°. Thus the permissive temperatures for groups I–V are 18°, 22°, 25°, 28°, and 30°, respectively. Note that the transition from one group to its neighbor is gradual, and within each group, the alleles are ordered top to bottom, with the bottom ones better able to support growth at a given temperature. Note also that all but one of the first-generation TS-intein alleles belong to groups IV and V. Thus, we have expanded the temperature range at which a TS intein can be used.
For convenience, we have given each TS-intein allele a three-digit identification number. The first digit represents the group to which a TS-intein allele belongs, while the last two digits represent its position in the group. For example, the ID number for S79, the 10th member of group II, is 210.
TS-intein splicing in Gal4p:
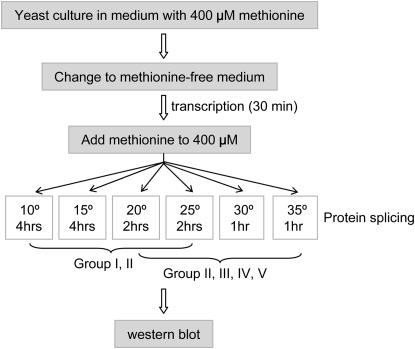
To determine whether the observed yeast growth profiles accurately reflected the splicing of the TS inteins, we transferred the Gal4-TS intein into the protein expression vector p416-Met25, which has a methionine-suppressive Met25 promoter (Mumberg et al. 1994), and assessed protein splicing directly by Western blotting (Figure 4 and Figure 5). As outlined in Figure 4, yeast carrying p416-Met25-Gal4 inteins were grown in methionine-containing media to 0.6 OD, shifted to methionine-free media to induce transcription for a fixed period of time, and then separated into aliquots that were incubated at different temperatures. For group I TS inteins and the first eight alleles of group II, yeast were incubated at 10°, 15°, 20°, and 25°; for the last four alleles of group II and all of groups III, IV, and V, yeast were incubated at 20°, 25°, 30°, and 35°. As with any other chemical reaction, the intein splicing rate varies with temperature, so splicing was allowed to proceed for different times at different temperatures. After preliminary testing, we decided to use 4 hr at 10° and 15°, 2 hr at 20° and 25°, and 1 hr at 30° and 35°.
Figure 4.—
Protocol for protein expression and assessing splicing. Yeast was first grown in the presence of methionine to the desired log-phase density and then placed in methionine-free media to induce transcription. Half an hour later, methionine was added to stop transcription. The yeast cultures were then divided and incubated at various temperatures for protein splicing. The results were analyzed by Western blotting.
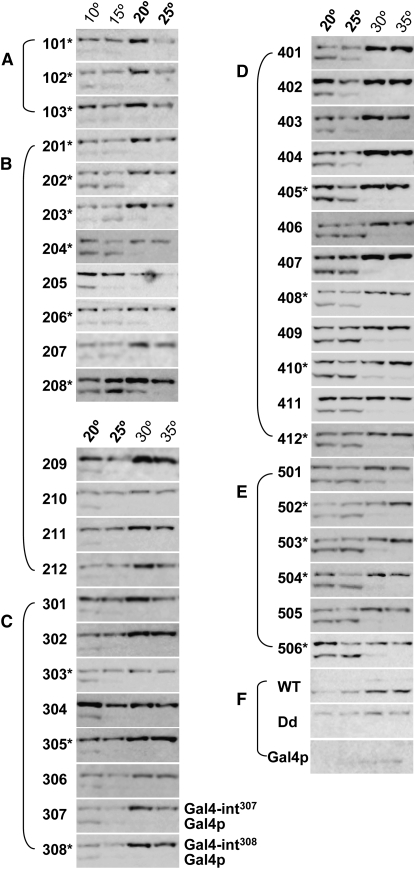
Figure 5.—
Western blot of Gal4-intein splicing. (A) Group I. (B) Group II. (C) Group III. (D) Group IV. (E) Group V. (F) Controls. Temperatures (20° and 25°) common for testing all TS-intein alleles are in boldface type. The top band in each panel is the unspliced precursor protein, while the bottom band is the spliced product Gal4p (labeled only for alleles 307 and 308). The alleles marked with an asterisk splice efficiently in both Gal4p and eGFP (Figure 6 and Figure S3) and are recommended for future use. The numbering and marking system used in this figure remains the same in the figures that follow, as well as in the figures in the supporting information. Note that the TS-intein alleles that enabled the host yeast to grow at higher temperatures also spliced at higher temperatures.
To stop splicing, yeast cells were lysed under denaturing conditions, and the resulting lysates were analyzed by Western blotting. Lysates of equal amounts of yeast were loaded in each lane. Note that the Western blots did not detect intein splicing per se, but a combination of intein splicing and several other processes, including the translation and degradation of Gal4-TS-intein RNA, as well as degradation of the fusion proteins and the splicing products (see Time-course analysis of eGFP-intein splicing below). Nonetheless, at the permissive temperature, spliced Gal4p is generated through protein splicing, and the concentration of the spliced product directly reflects how efficiently the functional host protein is produced from a particular Gal4-TS-intein fusion. Therefore, the relative levels of spliced Gal4p (bottom band in each panel in Figure 5) compared with its unspliced precursor protein (top band in each panel in Figure 5) were used to estimate how efficiently the TS-intein splices from its host protein.
As expected, all intein alleles spliced in a temperature-dependent manner (Figure 5). For intein alleles 101–202, splicing could be detected at 10° and 15° but not ≥20° (Figure 5, A and B). For alleles 203–207, weak splicing could be detected at 20° (Figure 5B). For alleles 208–308, splicing at 20° is obvious (Figure 5, B and C). For groups IV and V (Figure 5, D and E), significant splicing occurred at 25°. From allele 407 onward, weak splicing at 30° could be detected. No splicing was detected for inteinDd at any temperature, while inteinWT spliced at all temperatures (Figure 5F). The abilities of the TS-intein alleles with higher permissive temperatures to splice at higher temperatures demonstrate that the temperature-dependent yeast growth accurately reflects the splicing ability of the TS intein within the organism.
We noted that the temperatures at which the spliced form could be detected by Western blotting were lower than the permissive temperatures defined by the growth profiles. For example, for intein504 (Figure 5E), significant splicing was observed at 20° and 25°, but not at 30°, while the yeast grew at a temperature as high as 30° (Figure 3). This discrepancy may have two explanations. First, the splicing durations for the two assays are different—encompassing several days for the yeast growth assay, but only a few hours (varying from 1 to 4 hr) for the Western blot analysis. The other possible explanation is that there is a sensitivity difference between the two assay methods; much less of the functional Gal4p might be needed for growth than for visualization via Western blot detection.
TS-intein splicing in eGFP:
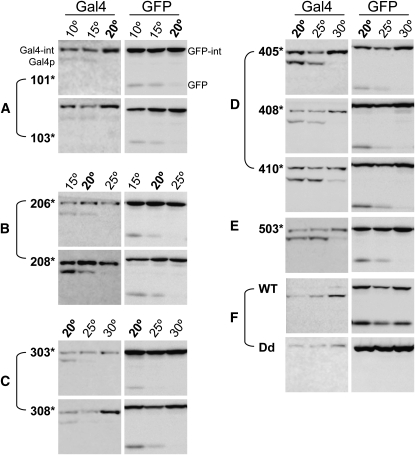
To determine the effects of host protein context on TS-intein splicing, we analyzed TS-intein splicing with enhanced green fluorescent protein (eGFP) (Figure 6 and Figure S3). The images of the Gal4-TS-intein alleles in Figure 6 were derived from the same Western blot as in Figure 5. Not surprisingly, TS inteins spliced in a temperature-sensitive manner within the new host protein eGFP. The same trend of splicing efficiency was observed in eGFP as was seen in Gal4p. It is remarkable that, for most intein alleles, the switching on/off temperatures were the same for Gal4p as for eGFP (Figure 6, A–E). There are, however, some exceptions. For example, eGFP-TS inteins 202, 207, and 411 barely spliced (Figure S3) while the corresponding Gal4 fusions spliced readily at their permissive temperatures (Figure 5). Interestingly, eGFP-TS intein103 spliced at temperatures as high as 22° (Figure S3), whereas we did not observe any Gal4-TS intein103 splicing at 20° or yeast growth at 22° (Figure 6A and Figure 3), suggesting that some alleles splice in a host-protein-dependent manner. Therefore, for each specific protein, one may need to test several TS-intein alleles. Fortunately, we have identified many alleles and the vast majority of these behaved identically in both host proteins.
Figure 6.—
Comparison of the splicing of selected TS-intein alleles in different host proteins. (A) Group I. (B) Group II. (C) Group III. (D) Group IV. (E) Group V. (F) Controls. WT: eGFP with wild-type intein inserted; Dd: eGFP with dead intein inserted. Temperature (20°) common for testing all TS-intein alleles is in boldface type. Note that TS-intein alleles spliced similarly in both host proteins. As in Gal4p, inteinWT splicing in eGFP was temperature independent, and inteinDd did not splice from its host protein eGFP.
Time-course analysis of eGFP-intein splicing:
To further compare the intein alleles, we analyzed the rate of intein splicing from eGFP and from BasGFP (Figure 7 and Figure S4). BasGFP is an eGFP fusion of bas1, a gene involved in histidine synthesis (Pinson et al. 1998). Several conclusions can be drawn from the time-course analyses of BasGFP-TS intein splicing:
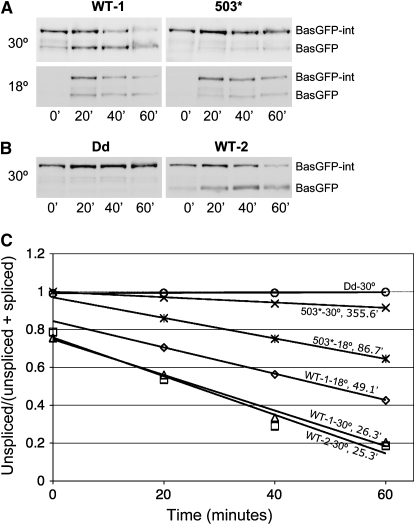
The rate of intein splicing changes with temperature. For example, the halftime (time at which the concentration of the unspliced product equals that of the spliced product) of inteinWT splicing was 49 min at 18° and <30 min at 30° (Figure 7, A and C).
Intein splicing is reproducible; the time courses of inteinWT splicing performed on two different dates are essentially the same (Figure 7C; also compare WT-1 at 30° in Figure 7A and WT-2 at 30° in Figure 7B).
TS inteins splice from BasGFP at a rate slower than inteinWT does. For example, at 18° the halftime was 49 min for inteinWT and 87 min for intein503 (Figure 7, A and C). The time-course analyses of eGFP-TS intein splicing show that different TS-intein alleles spliced at different rates and that significant splicing occurred within 1–2 hr at 18° for group II through group V TS inteins (Figure S4).
Figure 7.—
Time course of intein splicing in BasGFP. Western blot analysis with anti-GFP antibody of proteins from yeast carrying a BasGFP intein incubated for the indicated periods after stopping transcription induction at 18° or 30°. (A) Time courses of the splicing of inteinWT and intein503. At 30°, inteinWT (top left) spliced rapidly, while intein503 (top right) barely spliced. At 18°, intein503 (bottom right) spliced at a reasonable speed but was slower to splice than inteinWT (bottom left). (B) Time courses of the splicing of inteinWT and inteinDd. Dead intein did not splice from its host protein. Note that the time courses of the splicing of a BasGFP-inteinWT at 30° done on 2 different days (top left in A and right in B) were very similar, indicating that the splicing reaction is reproducible. (C) Quantification of BasGFP-intein splicing. The y-axis is the ratio of the amount of unspliced BasGFP intein to the total amount of BasGFP and BasGFP intein. The x-axis is the time (in minutes) when yeast cells were harvested. The moment at which transcription induction was stopped was time zero. The calculated, pseudo-first order, splicing halftime is presented following the temperature of splicing and the name of each intein allele.
As previously mentioned, the Western blot assays assess the aggregate effects of protein splicing, RNA turnover, translation, and protein degradation. The total effect of RNA turnover, translation, and unspliced protein degradation can be appreciated by examining the protein levels of Bas-GFP-inteinDd, which is unable to splice (Figure 7B, left). The decrease in the levels of BasGFP, after the initial increase, even with the continuous addition of BasGFP resulting from the ongoing splicing, is the result of the turnover of the spliced product (Figure 7B, right; compare lane 3 to lane 4).
Consistent with the intein splicing characteristics from within Gal4p and eGFP, inteinDd did not splice from its host protein BasGFP (Figure 7, B and C), and inteinWT spliced from BasGFP at both 18° and 30° (Figure 7, A–C). And while intein503 spliced from BasGFP readily at 18°, it did so only slightly at 30° (Figure 7, A and C).
DISCUSSION
Conventional methods for producing TS alleles use chemical mutagens to generate random mutations in a host organism. Following mutagenesis, a large number of progeny are screened for a desired TS phenotype, and the gene responsible for the phenotype is identified. This approach has a number of drawbacks for functional genomics in that the process is labor intensive, and not all proteins can be mutated to temperature sensitivity (Suzuki et al. 1971; Harris and Pringle 1991). In addition, this approach is feasible only in organisms in which a large number of progeny can be screened.
A number of in vitro mutagenesis approaches have been developed to generate TS alleles for a particular gene of interest. One approach is to clone the gene of interest and randomly mutagenize it via low-fidelity PCR. The mutagenized DNA is then introduced into a host organism and screened in the context of a null background. The frequency of generating TS alleles using this approach is quite low and therefore also requires screening a large number of progeny. A more direct technique is to replace charged amino acid clusters with alanines (Wertman et al. 1992). Applying this approach to the actin gene of S. cerevisiae resulted in 44% of the generated mutations being TS in vivo. In contrast, another approach modifies buried hydrophobic cores within a protein (Chakshusmathi et al. 2004). The second method was applied to the yeast GAL4 gene, and several TS alleles were generated. In comparison, Chakshusmathi et al. (2004) also mutagenized GAL4 via low-fidelity PCR. Over 20,000 transformants were obtained, but none was TS. Thus the efficiency of directly mutating charged amino acid clusters or hydrophobic cores to yield a TS allele is much greater than that of random mutagenesis. However, it is not practical to generate TS mutations for a large number of proteins by individually cloning and mutagenizing each gene.
A more general method for creating TS alleles for genomewide application in S. cerevisiae uses a degron (Dohmen et al. 1994; Kanemaki et al. 2003; Dohmen and Varshavsky 2005). At 37°, the degron, and whatever it is fused to, are targeted for degradation via a ubiquitin-mediated “N-end rule” pathway that recognizes aberrantly folded proteins. At permissive temperatures, the degron does not activate the N-end rule pathway, and the fusion protein is not targeted for degradation. Kanemaki et al. (2003) tagged 103 essential yeast genes with the degron, of which 60% behaved in a temperature-sensitive manner. Application of this approach to multicellular eukaryotes has met with limited success (Levy et al. 1999; Lindner et al. 2002). Furthermore, the inactivating temperature, 37°, is too high for many organisms, including D. melanogaster, to survive long enough for phenotypic analysis.
Temperature-sensitive activities of the host proteins of GyrA intein and recA intein were reported previously (Derbyshire et al. 1997; Adam and Perler 2002). The GyrA intein variants represent true splicing mutants; however, the splicing of the GyrA intein requires specific residues at the N extein, making it unsuitable as a general tool for controlling the activities of other proteins (Telenti et al. 1997; Southworth et al. 1999). With respect to the recA intein variants, it is unclear if they are true TS alleles (Hiraga et al. 2005). We chose Sce VMA to make our TS-intein switch because Sce VMA splices itself efficiently from many ectopic locations (Hirata et al. 1990; Kane et al. 1990; Chong et al. 1998). As a proof of principle, TS inteins have been successfully used in bacteria, yeast, D. melanogaster, and zebrafish (Zeidler et al. 2004; Liang et al. 2007; S. Holley, personal communication) to conditionally control protein activities. However, the first-generation TS inteins were limited in that the number of alleles was small, and most of them had relatively high permissive temperatures.
We have generated a large collection of new TS-intein alleles with a broad range of temperatures at which the TS intein can be used. The classification, splicing analysis, and studies of the splicing kinetics of both the old and the newly generated TS-intein alleles have provided a guideline for one to choose the most appropriate intein switch. Sce VMA splices efficiently when inserted directly upstream of a cysteine residue in the host protein. Thus, these intein switches can be used with wild type, as well as constitutively active or dominant-negative forms of a protein as long as they contain cysteine residues or a cysteine residue can be added without affecting the function of the protein. Researchers will be able to choose intein(s) that function at an optimal temperature for their specific application. Note that each target protein folds in a unique manner, and thus the specific intein insertion site may affect subsequent excision. It might therefore be necessary to try different insertion sites to generate a TS-intein allele that excises properly. This can also be addressed by inserting different intein alleles with the same permissive temperature and by assessing for a TS phenotype because some may excise more favorably than others from a particular insertion site. On a related note, it is possible that certain regions of a specific host protein can tolerate a large insertion; consequently, at the nonpermissive temperature, the inserted TS intein may generate only a hypomorphic allele or may have no effect on the activity of the host protein. Thus it may be necessary to assess multiple insertion sites and different intein alleles to generate a TS protein that is nonfunctional at the nonpermissive temperature. Our collection of a family of TS-intein switches that function at distinct temperatures constitutes a new toolbox for the generation of TS mutations tailored either for the growth requirements of one's particular organism or for experimental conditions.
Acknowledgments
We thank F. Winston (Harvard Medical School) and B. Daignan-Fornier (Institut de Biochimie et Genetique Cellulaires) for yeast strains; K. Cone, K. Bennett, B. McClure, T. Zars, and S. Ong at the University of Missouri, as well as R. Binari at Harvard Medical School, for constructive discussions. This project was funded by start-up funds provided by the University of Missouri and a grant from the National Science Foundation (MCB-0641309).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.104794/DC1.
References
- Adam, E., and F. B. Perler, 2002. Development of a positive genetic selection system for inhibition of protein splicing using mycobacterial inteins in Escherichia coli DNA gyrase subunit A. J. Mol. Microbiol. Biotechnol. 4: 479–487. [PubMed] [Google Scholar]
- Anraku, Y., R. Mizutani and Y. Satow, 2005. Protein splicing: its discovery and structural insight into novel chemical mechanisms. IUBMB Life 57: 563–574. [DOI] [PubMed] [Google Scholar]
- Buskirk, A. R., Y. C. Ong, Z. J. Gartner and D. R. Liu, 2004. Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. USA 101: 10505–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, R. C., and G. F. Joyce, 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2: 28–33. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi, G., K. Mondal, G. S. Lakshmi, G. Singh, A. Roy et al., 2004. Design of temperature-sensitive mutants solely from amino acid sequence. Proc. Natl. Acad. Sci. USA 101: 7925–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, S., K. S. Williams, C. Wotkowicz and M. Q. Xu, 1998. Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J. Biol. Chem. 273: 10567–10577. [DOI] [PubMed] [Google Scholar]
- Costa, R. M., C. Drew and A. J. Silva, 2005. Notch to remember. Trends Neurosci. 28: 429–435. [DOI] [PubMed] [Google Scholar]
- Derbyshire, V., D. W. Wood, W. Wu, J. T. Dansereau, J. Z. Dalgaard et al., 1997. Genetic definition of a protein-splicing domain: functional mini-inteins support structure predictions and a model for intein evolution. Proc. Natl. Acad. Sci USA 94: 11466–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R. J., and A. Varshavsky, 2005. Heat-inducible degron and the making of conditional mutants. Methods Enzymol. 399: 799–822. [DOI] [PubMed] [Google Scholar]
- Dohmen, R. J., P. Wu and A. Varshavsky, 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263: 1273–1276. [DOI] [PubMed] [Google Scholar]
- Evans, T. J. T., and M. Q. Xu, 2002. Mechanistic and kinetic considerations of protein splicing. Chem. Rev. 102: 4869–4884. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., and J. R. Pringle, 1991. Genetic analysis of Saccharomyces cerevisiae chromosome I: on the role of mutagen specificity in delimiting the set of genes identifiable using temperature-sensitive-lethal mutations. Genetics 127: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga, K., V. Derbyshire, J. T. Dansereau, P. Van Roey and M. Belfort, 2005. Minimization and stabilization of the Mycobacterium tuberculosis recA intein. J. Mol. Biol. 354: 916–926. [DOI] [PubMed] [Google Scholar]
- Hirata, R., Y. Ohsumk, A. Nakano, H. Kawasaki, K. Suzuki et al., 1990. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265: 6726–6733. [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Kane, P. M., C. T. Yamashiro, D. F. Wolczyk, N. Neff, M. Goebl et al., 1990. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science 250: 651–657. [DOI] [PubMed] [Google Scholar]
- Kanemaki, M., A. Sanchez-Diaz, A. Gambus and K. Labib, 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–724. [DOI] [PubMed] [Google Scholar]
- Levy, F., J. A. Johnston and A. Varshavsky, 1999. Analysis of a conditional degradation signal in yeast and mammalian cells. Eur. J. Biochem. 259: 244–252. [DOI] [PubMed] [Google Scholar]
- Liang, R., X. Liu, J. Liu, Q. Ren, P. Liang et al., 2007. A T7-expression system under temperature control could create temperature-sensitive phenotype of target gene in Escherichia coli. J. Microbiol. Methods 68: 497–506. [DOI] [PubMed] [Google Scholar]
- Lindner, K., J. Gregan, S. Montgomery and S. E. Kearsey, 2002. Essential role of MCM proteins in premeiotic DNA replication. Mol. Biol. Cell 13: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos, G. L., and G. M. Rubin, 1996. The role of the genome project in determining gene function: insights from model organisms. Cell 86: 521–529. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22: 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler, F. B., 2005. Protein splicing mechanisms and applications. IUBMB Life 57: 469–476. [DOI] [PubMed] [Google Scholar]
- Pinson, B., I. Sagot, F. Borne, O. S. Gabrielsen and B. Daignan-Fornier, 1998. Mutations in the yeast Myb-like protein Bas1p resulting in discrimination between promoters in vivo but not in vitro. Nucleic Acids Res. 26: 3977–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presente, A., A. Andres and J. S. Nye, 2001. Requirement of Notch in adulthood for neurological function and longevity. Neuroreport 12: 3321–3325. [DOI] [PubMed] [Google Scholar]
- Pringle, J. R., 1975. Induction, selection, and experimental uses of temperature-sensitive and other conditional mutants of yeast. Methods Cell Biol. 12: 233–272. [DOI] [PubMed] [Google Scholar]
- Raymond, C. K., T. A. Pownder and S. L. Sexson, 1999. General method for plasmid construction using homologous recombination. Biotechniques 26: 134–138, 140–131. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsh and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, NY.
- Shellenbarger, D. L., and J. D. Mohler, 1978. Temperature-sensitive periods and autonomy of pleiotropic effects of l(1)Nts1, a conditional notch lethal in Drosophila. Dev. Biol. 62: 432–446. [DOI] [PubMed] [Google Scholar]
- Southworth, M. W., K. Amaya, T. C. Evans, M. Q. Xu and F. B. Perler, 1999. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques 27: 110–114, 116, 118–120. [DOI] [PubMed] [Google Scholar]
- Suzuki, D. T., T. Grigliatti and R. Williamson, 1971. Temperature-sensitive mutations in Drosophila melanogaster. VII. A mutation (para-ts) causing reversible adult paralysis. Proc. Natl. Acad. Sci. USA 68: 890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti, A., M. Southworth, F. Alcaide, S. Daugelat, W. R. Jacobs, Jr. et al., 1997. The Mycobacterium xenopi GyrA protein splicing element: characterization of a minimal intein. J. Bacteriol. 179: 6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung and P. Philippsen, 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13: 1065–1075. [DOI] [PubMed] [Google Scholar]
- Wertman, K. F., D. G. Drubin and D. Botstein, 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132: 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler, M. P., C. Tan, Y. Bellaiche, S. Cherry, S. Hader et al., 2004. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 22: 871–876. [DOI] [PubMed] [Google Scholar]