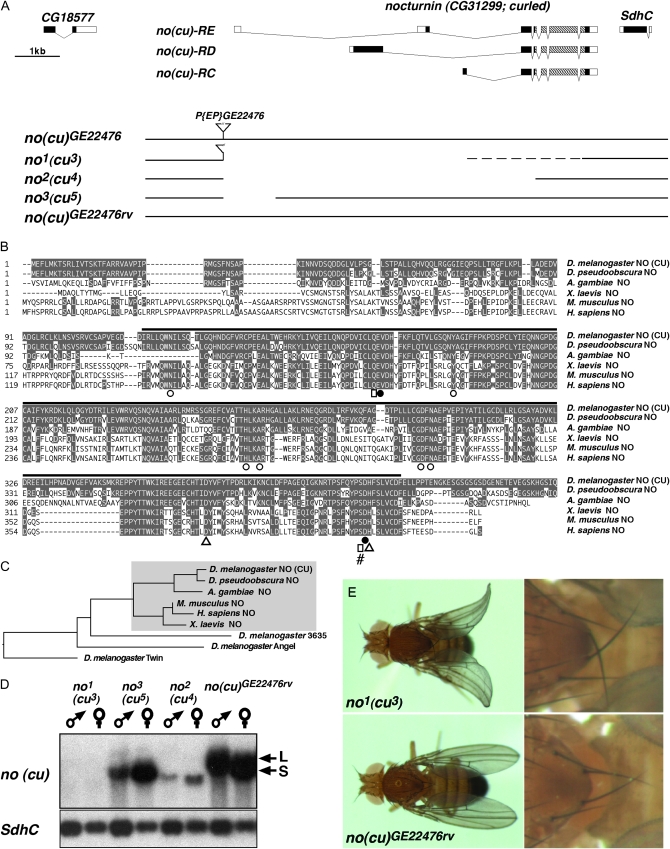
Figure 1.—
The Drosophila nocturnin (curled) gene: gene locus organization, deletion mutants, and phylogeny of Nocturnin proteins. (A) Organization of nocturnin (curled) gene locus with transcript isoforms no(cu)-RC, no(cu)-RD, and no(cu)-RE relative to the flanking genes CG18577 and SdhC at 86D7 on chromosome 3R. [Note: Coding parts of exons are marked by black, noncoding parts by white boxes; hatched boxes, Mg2+-dependent endonuclease-like domain (pfam03372); no(cu)-RC, -RD, -RE are the only no(cu) transcripts annotated in FlyBase r5.17.] Transposon integration line P{EP}no(cu)GE22476 was used to generate nocturnin (curled) deletion mutants no1 (cu3), no2 (cu4), and no3 (cu5) as well as genetically matched no+ (cu+) control line no(cu)GE22476rv. (B) Sequence alignment of vertebrate und invertebrate Nocturnin (NO) protein family members proves strong evolutionary conservation of amino acids involved in the catalytic function of yCCR4-related family proteins (amino acids identical to D. melanogaster NO are shaded in gray). Residues essential for the Mg2+-dependent endonuclease-like domain function are indicated as in Dupressoir et al. (2001): ▵, catalytic residue; □, residues involved in orientation and stabilization of catalytic residues; ○, for phosphate binding, and •, for Mg2+ binding residues; #, position of stop in nostop12.2. The black bar illustrates the extent of the pfam03372 domain. (C) Phylogenetic tree analysis showing that Drosophila NO(CU) is the Nocturnin ortholog (Nocturnin subfamily shaded gray) among the four yCCR4-related proteins of the fly. (D) Northern blot analysis of adult flies shows nocturnin (curled) small S and large L transcript populations in no(cu)GE22476rv controls and identifies no1 (cu3) as transcript null mutation. No3 (cu5) specifically lacks no(cu) L transcripts, while no2 (cu4) expresses a low abundance transcript slightly shorter than the no(cu) S transcript of control flies. SdhC transcript is unaffected in all no(cu) mutants. (E) Distally upward bent wing (left panel) and proximally crossed posterior scutellar bristle (right panel) phenotypes of no1 (cu3) mutants compared to no(cu)GE22476rv controls.