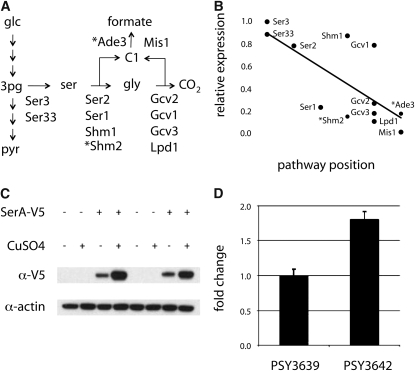
Figure 6.—
Flux to formic acid is controlled by PGDH and the coordinated expression of all pathway enzymes. (A) The subnetwork of serine–glycine–formate biosynthesis with reaction arrows that represent composites of several biochemical interconversions labeled with relevant enzymes. The cytoplasmic enzymes involved in converting serine into formic acid (Shm2 and Ade3) are denoted with asterisks. (B) Differential expression of enzymes in A were normalized to Ser3 and plotted according to their pathway position. Multisite modulation for enzymes in the formic acid biosynthetic pathway is indicated by linear regression (solid line, R2 = 0.52) and corrected Spearman's ranked order correlation (ρ = −0.72, P = 0.02). (C) Levels of the SerA fusion protein (relative to actin) were unaffected by genetic background and induction conditions. (D) Expressing SerA in PSY3642 caused a significant increase in formic acid secretion under inducing conditions. Each transformed strain was normalized to the empty vector control plasmid pRS410a. Data in D represent the average of three biological replicates ± SD. We assessed statistical significance using a paired two-tailed t-test (P = 0.002).