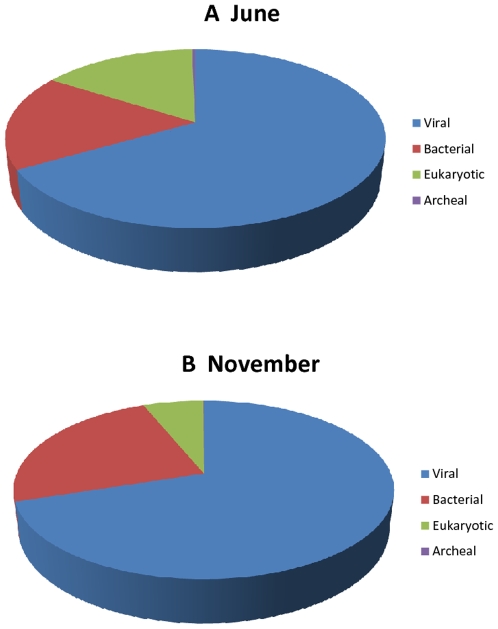
Figure 1. Taxonomic classification of assemblies.
Assemblies were classified based on comparison to the CAMERA database using the BLASTX algorithm and an e-value of 1×10e-5 or lower. Sequences in assemblies without significant matches to existing protein sequences (e-value>1E-5) were classified as “Unknown”. The remaining sequences were classified based on best BLASTX hits for their assemblies. Of the “known” sequences, 67% of the November sample and 70% of the June sample had homology to published viral sequences.