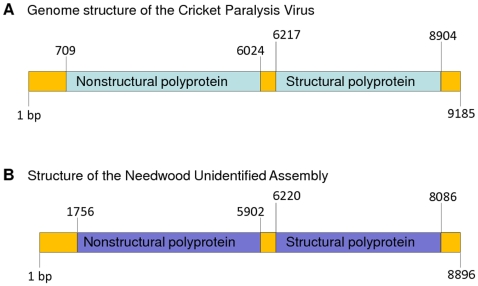
Figure 6. Genomic organization of a novel paralysis virus.
(A) Genomic organization of cricket paralysis virus. (B) Genomic organization of the putative novel paralysis virus identified in Lake Needwood. Two contiguous sequences with sizes of 6000 and 1500 nucleotides assembled from combined June and November reads exhibited significant homology with the cricket paralysis virus. Using targeting amplification a DNA fragment of 700 nt was amplified, sequenced and used to link the two contigs thus generating a consensus sequence of 8086 nt. Using BLASTX we mapped the boundaries of the two (non structural and structural) polyproteins. (C) Phylogenetic analysis of the Lake Needwood virus assembly with homologous paralysis viruses. A region (containing ∼600 amino acids residues) of the replicase polyprotein was used for phylogenetic analysis after multiple sequences alignment using ClustalX with default parameter settings as described in Materials and Methods. Consensus tree bootstrapping was performed with Geneious 4.0.4 using the neighbor-joining method and 1,000 samples.