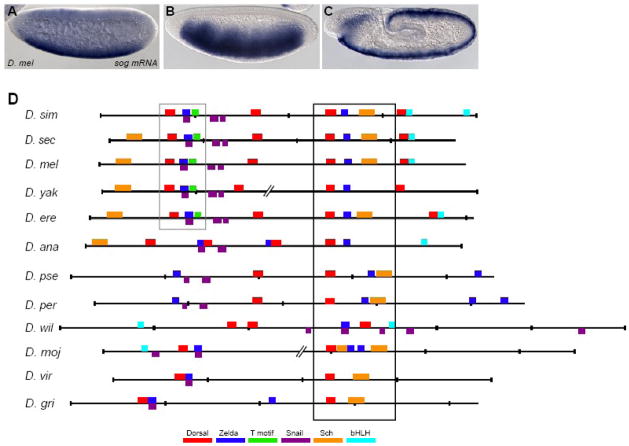
Fig. 2. Alignment of putative sog cis-regulatory elements from other Drosophilids reveals conservation and turnover of binding sites.
(A–C) sog expression is dynamic. Endogenous expression of the sog gene detected by in situ hybridization using a riboprobe within embryos of nuclear cycle ~11 (A), cycle 14/stage 5 (B), and during germ-band elongation after gastrulation (C). (D) Shown are the predicted binding sites for transcription factors and over-represented motifs that are associated with neurogenic ectoderm patterning within the sog cis-regulatory modules identified from 12 Drosophilid species. Position weight matrices (PWMs) were used to find putative transcription factor binding sites using the program Cartwheel (Brown et al., 2005). Alignments were generated on the UCSC genome browser webpage (http://genome.ucsc.edu). Cartoons were generated by Cartwheel and then colored according to the key. Gaps in the alignments, shown as broken lines, were introduced to help visualize conservation. Box domains represents well-conserved region of sog cis-regulatory element. The sites boxed in black are located in the most well-conserved region, and the sites boxed in grey are the second most well-conserved region. Note that closely associated Dorsal and Zelda binding sites are present in all of the alignments, though the location of these sites within the sequence can vary. Full alignment can be viewed in Supplemental Fig. 3.