Figure 6. The Ska1 complex coated microspheres display microtubule depolymerization driven motility.
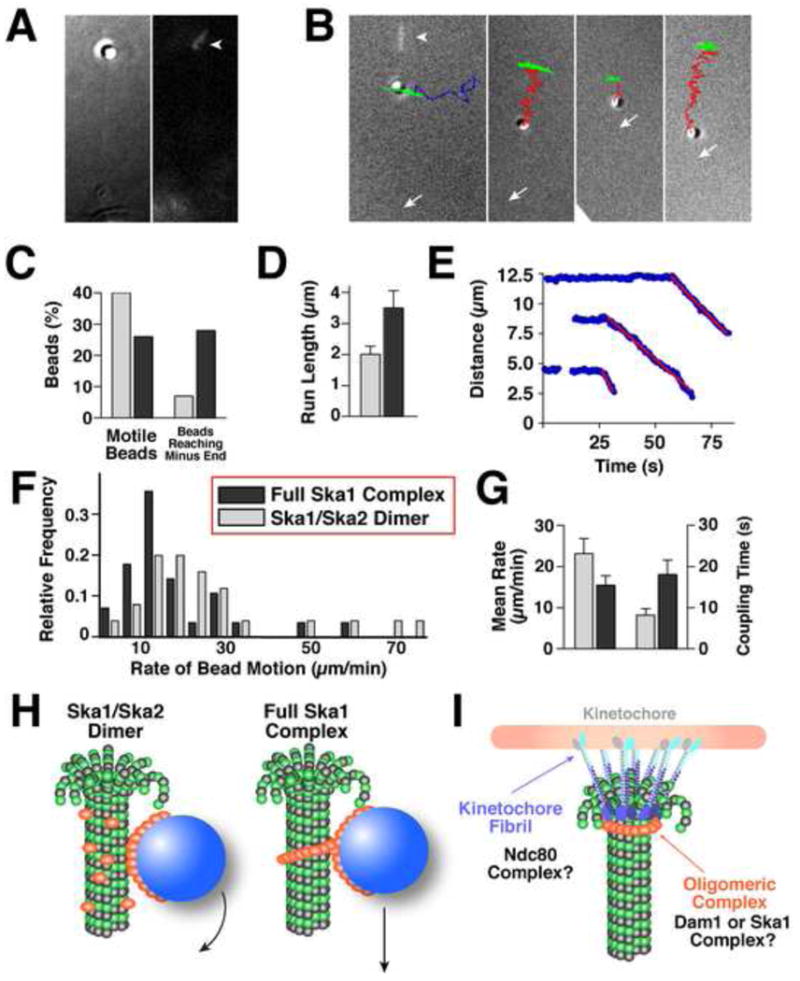
(A) DIC image (left panel) of a microtubule-bound bead with a corresponding fluorescent image (right panel) showing the stabilized microtubule cap (arrowhead). (B) Example trajectories of moving Ska1 complex-coated beads overlayed with the terminal corresponding DIC image. The trajectory of a bead that is stably attached to the wall of a static microtubule polymer (green) resembles a short arc, because the unattached microtubule plus end swings randomly as the overall microtubule length remains constant. Bead detachment from a microtubule is evidenced by a change of this swinging motion into a random “walk” (blue trajectory). Microtubule depolymerization-driven bead motions (red trajectories) are linearly directed and the amplitude of their swinging motions diminishes as the microtubule shortens. Arrows indicate positions of the coverslip-attached microtubule minus ends. First panel additionally shows an overlayed fluorescent image of the rhodamine-labeled microtubule cap before photo-ablation (arrowhead). (C) Graph showing the percent of beads that moved >0.5 μm and percent of moving beads that reached the microtubule end (full Ska1 complex, n = 112; Ska1/Ska2 dimer, n = 68). (D) Graph showing the mean distances traveled (+/- standard error of the mean - SEM) for Ska1 complex and Ska1/Ska2 dimer coated beads. (E) Graph showing the relative distance between the microsphere and microtubule minus end for the processively moving beads shown in (B). Red lines are the best linear fit for the descending segments of these curves. (F) Histogram distribution showing the motility rates of the Ska1/Ska2 dimer (n = 25) and full Ska1 complex (n = 28) coated microspheres. (G) Graph showing the average rates of bead motions and the time of bead attachment to the shortening microtubule end. Error bars are SEM. (H-I) Speculative model for Ska1 complex assembly and function. (H) The bead motions of the Ska1/Ska2 dimer are similar to Dam1 complex that does not form a ring-like structure, and may be due to the rolling of the beads. The full Ska1 complex moves at a similar rate to ring-like Dam1 complex. In this model, the Ska1 complex is shown as an oligomeric structure on microtubules. The oligomeric nature of Ska1 is based on the strong cooperativity in microtubule binding, the structures visualized on microtubules by EM, and the movement of the Ska1 along microtubules at a rate that suggests it creates an impediment to microtubule depolymerization. (I) Distinct microtubule coupling activities at the kinetochore include a fibril-like structure such as the Ndc80 complex and an oligomeric assembly such as the Dam1 or Ska1 complex.
