Abstract
The misfolding of nascent proteins, or the unfolding of proteins after synthesis is complete, can occur in response to numerous environmental stresses, or as a result of mutations that de-stabilize protein structure. Cells have developed elaborate protein quality control systems that recognize improperly folded proteins and either refold them, or facilitate their degradation. One such quality control system is the unfolded protein response, or the UPR. The UPR is a highly conserved signal transduction system that is activated when cells are subjected to conditions that alter the endoplasmic reticulum (ER) in ways that impair the folding of nascent proteins in this organelle. Recent observations indicate that in the heart, the UPR is activated during acute stresses, including ischemia/reperfusion, as well as upon longer-term stresses that lead to cardiac hypertrophy and heart failure. Moreover, certain aspects of the UPR are activated during, and are required for proper heart development. This review summarizes recent studies of the UPR in the heart, focusing on the possible roles of the UPR in contributing to, or protecting from ischemia/reperfusion damage.
Introduction
The function of most proteins depends on their 3-dimensional conformation, which requires correct folding. The bulk of cellular protein synthesis takes place on cytosolic free ribosomes. However, depending on the cell type, up to 35% of protein synthesis is in the rough endoplasmic reticulum (ER), which is where all secreted proteins, as well as proteins that are targeted to membranes and organelles are synthesized [1]. Under optimal conditions, numerous chaperones and other proteins and factors, ensure efficient nascent protein folding; however, perturbation of folding machinery components decreases protein folding efficiency. Initially, the accumulation of misfolded proteins triggers biochemical events designed to augment protein folding capacity. However, the continued accumulation of terminally misfolded proteins leads to their degradation by a well studied protein quality control system, involving recognition, ubiquitination and degradation by proteasomes and, eventually, to cell death. Protein quality control systems are located in several cell locations, including the cytosol and the ER; several reviews of the cytosolic protein quality control system in the heart have recently appeared [2, 3]. Accordingly, this review focuses on ER-associated protein quality control system, also known as the unfolded protein response (UPR).
The Unfolded Protein Response
The unfolded protein response (UPR) is a conserved signal transduction system that is activated by stresses that impact the efficiency of protein folding in the rough ER [4-7]. Optimal protein folding in the ER depends on maintaining the the proper balance of numerous components in the ER that contribute to folding of proteins during synthesis. For example, the correct ER redox status is required for protein disulfide bond formation, which is an oxygen-requiring process. Also, maintenance of suitable levels of protein glycosylation substrates, as well as sufficient glycosylation enzymatic machinery, are required, since most proteins made in the ER are glycosylated. Finally, ER calcium and ER resident chaperones must be present at the levels that facilitate optimal folding of nascent ER proteins.
Experimentally, the ER environment can be perturbed by substances, such as dithiothrietol, thapsigargin, or tunicamycin, which alter redox status, calcium levels and protein glycosylation in the ER, respectively [4, 8-10]. When cells are treated with one of these compounds, or if they are starved of glucose and oxygen, the latter of which mimics ischemia, ER protein folding is impaired, and the accumulation of mis-folded, dysfunctional proteins signals the initiation of ER stress [11].
ER stress is initially sensed by the 3 ER-transmembrane proteins, protein kinase R-like ER kinase (PERK) [12], activating transcription factor 6 (ATF6) [13, 14] and inositol-requiring enzyme-1 (IRE-1) [15, 16], which serve as the primary proximal effectors of the UPR. Numerous studies in yeast, as well as mammalian cell lines and a few tissues have contributed considerably to our understanding of the mechanisms of action of each of these effectors. Several excellent reviews report on the results of these studies [17]; accordingly, the following is a summary of the mechanisms by which these effectors sense ER stress and mediate downstream signals.
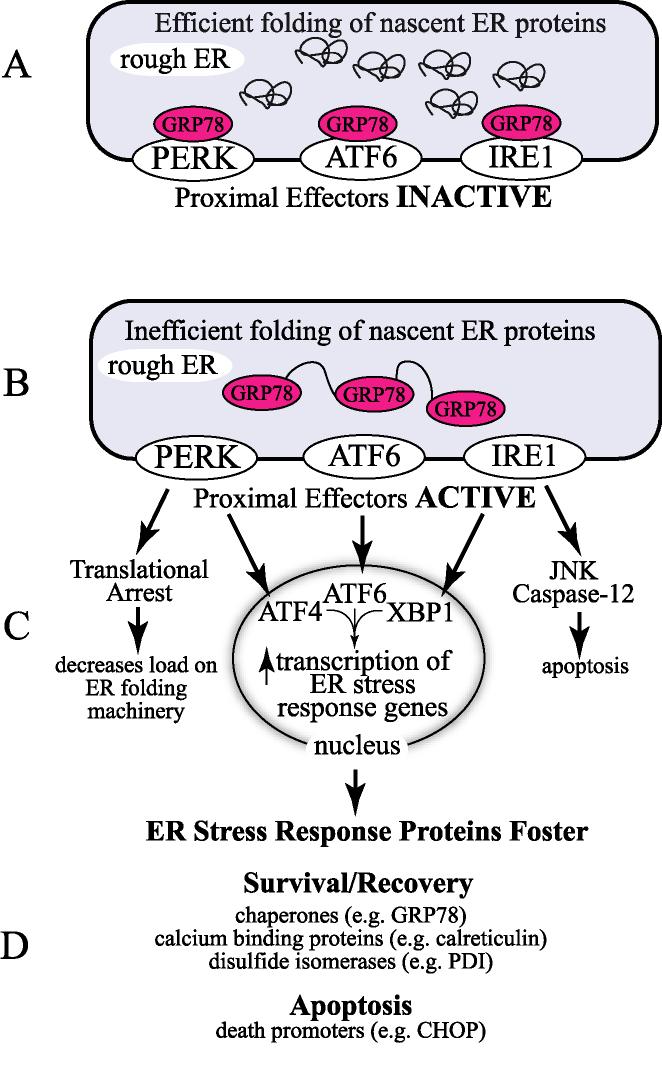
When ER protein folding is functioning efficiently, the ER luminal domains of PERK, ATF6 and IRE-1 are bound to the abundant ER-resident chaperone, glucose-regulated protein 78 (GRP78). Under these conditions, these proximal effectors of the UPR are inactive (Fig. 1A). However, when ER protein folding is disrupted, and misfolded proteins begin to accumulate, GRP78 translocates from PERK, IRE-1 and ATF6 to the misfolded proteins in an apparent effort to aid in folding [18-22]. This translocation of GRP78 leads to the activation of all 3 proximal effectors of the UPR (Fig. 1B), and is considered one of the earliest signs of ER stress. Once activated, the 3 proximal effectors of the UPR mediate the downstream, or distal effects of ER stress.
Figure 1.
The Unfolded Protein Response:
Panel A- Under non-stressed conditions, proteins that are synthesized in the rough ER are efficiently folded. The ER-resident molecular chaperone, glucose-regulated protein-78 (GRP78), is associated with the luminal domains of the 3 proximal effectors of the unfolded protein response (UPR), PKR-like ER kinase (PERK), inositol-required enzyme-1 (IRE-1) and activating transcription factor 6 (ATF6). Under these conditions, these 3 UPR effectors are inactive in terms of activating downstream effects of the UPR.
Panel B- Stresses that perturb the redox status of the ER lumen, alter ER calcium levels, or disrupt the ER protein glycostylation machinery, result in the accumulation of mis-folded, dysfunctional proteins in the ER lumen, which initiates ER stress. Upon ER stress, GRP78 translocates from the luminal domains of PERK, ATF6 and IRE-1 to the misfolded proteins in the ER lumen in an effort to assist in folding. Under these conditions, PERK, ATF6 and IRE-1 and ATF6 are activated.
Panel C- The PERK, ATF6 and IRE-1 branches of the UPR each mediate the transcriptional induction of ER stress response genes via the transcription factors, ATF4, ATF6 and XBP1, respectively. In addition, PERK can mediate global translational arrest by phosphorylation eIf2α, and IRE-1 can mediate the activation of JNK and caspase-12. Translational arrest decreases the workload on the ER, allowing time for recovery; however, the mRNAs encoded by many ER stress response genes have evolved structural features that allow them to escape this translational arrest, which is important for the up-regulation of ER stress response proteins upon acute stress. The JNK and caspase-12 branches of the UPR contribute to programmed cell death that takes place upon chronic ER stress. Many ER stress response genes encode ER-targeted chaperones, and other ER proteins that are designed to stabilize and/or re-establish an ER luminal environment that is suitable for nascent ER protein folding, facilitating cell survival and recovery from acute ER stress (Survival/Recovery). However, if the stress is not resolved, ER stress response genes induced upon chronic stress lead to apoptotic cell death (Apoptosis).
Proximal Effectors of ER Stress (PERK, IRE-1 and ATF6)
PERK
PERK is a transmembrane ER protein; upon ER stress, and the relocation of GRP78 from the luminal domain of PERK to misfolded proteins, and the subsequent homodimerization of PERK, lead to trans-autophosphorylation, much like activated growth factor receptors. This autophosphorylation activates PERK, further, which phosphorylates the ribosomal protein, eIF2α. Phosphorylation of eIF2α decreases its efficiency as an initiator of translation, which leads to decreased translation of most cellular mRNAs (Fig. 1C; translation arrest) [18]. This translational arrest, which is transient, reduces the protein synthesis load in the ER, facilitating recovery of ER homeostasis and the re-establishment of efficient ER protein folding [23]. Although most mRNAs are inefficiently translated upon PERK activation, paradoxically, the mRNA that encodes activator of transcription factor 4 (ATF4) is translated more efficiently when eIF2α is phosphorylated. This leads to increased levels of ATF4, which serves important roles as a transcriptional inducer of a certain ER stress response genes, such as those that encode amino acid transporters, which assist in recovery from the stress (Fig. 1C; ATF4).
IRE-1
Much like PERK, IRE-1 is an ER transmembrane protein, which, upon ER stress, forms homodimers which facilitate trans-autophosphorylation. However, in contrast to PERK, upon ER stress, IRE-1 exhibits a novel endoribonuclease activity, which splices the mRNA that encodes active x-box binding protein-1, (XBP1). This unusual splicing event, which takes place in the cytosol, generates a transcript with a new open reading frame that encodes the expression of an active form of XBP1, a basic leucine-zipper (bZip) transcription factor that induces numerous ER stress response genes (Fig. 1C; IRE1 XBP1) [21, 24].
ATF6
Like PERK and IRE-1, ATF6 is an ER transmembrane protein that also exists as a dimer in association with GRP78 under non-stressed conditions. In further comparison, upon ER stress GRP78 dissociates from the ER-luminal domain of ATF6. However, in contrast to the other two effectors, which remain associated with the ER, ATF6 re-locates to the Golgi, where two proteases, site-1 and site-2 proteases, cleave it in, or near the transmembrane region. After these cleavage events, the cytosolic region of ATF6, which has several putative nuclear localization signals, translocates to the nucleus where it can form homodimers or heterodimers with a small group of b-Zip transcription factors, which includes XBP1, leading to the transcriptional regulation of ER stress response genes (Fig. 1C; ATF6) [25, 26]. A second isoform of ATF6, ATF6β [27], as well as other ATF6-related proteins, such as Oasis [28], Luman [29, 30], CREB4 [31], CREB-H [31] and BBF2H7 [32], have since been discovered. Like ATF6, all of these isoforms are ER trans-membrane proteins that are cleaved and translocate to the nucleus upon ER stress. Moreover, in theory, each has the ability to dimerize with other members of this b-Zip transcription factor family. Although the exact roles of these ATF6 isoforms are not completely known, it appears as though ATF6β has less ability to induce ER stress response genes than ATF6 [33], and may even serve as an inhibitor of ATF6 [34, 35]. Additionally, some of the other isoforms exhibit tissue-restricted expression patterns, implying that they may contribute to mediating ER stress in a cell type specific manner.
Distal Effectors of the UPR
Many of the XBP1 and ATF6 inducible genes that have been characterized to date can be induced by either transcription factor. Thus, there is a great deal of redundancy between these two UPR pathways, although the reasons for this redundancy are not yet clear. However, the recent development of ATF6 knock out mice has clarified the existence of numerous ER stress response genes that are dependent upon ATF6 for maximal induction during ER stress [33]. Most of the genes induced by XBP1 and/or ATF6 encode mRNAs with structural features allowing them to escape PERK-mediated translational arrest. Genes induced upon acute ER stress encode proteins that improve the folding of nascent proteins in the ER lumen and facilitate the degradation of dysfunctional misfolded proteins. The degradation of terminally misfolded ER proteins is performed by ER associated protein degradation, or ERAD. ERAD is a complex process involving the recognition of misfolded ER proteins, followed by the retrotranslocation of these proteins across the ER membrane to the cytosolic face of the ER. Protein degradation machinery, located on the cytosolic face of the ER, is dedicated to the ubiquitination and proteasome-mediated degradation of terminally misfolded ER proteins [36-38]. Together, the ER-luminal and ERAD-associated proteins function to resolve the ER stress, fostering the recovery of efficient ER protein folding and cell survival (Fig. 1D; Survival/Recovery).
If the UPR signals activated in the early phases of ER stress are not sufficient to resolve the stress, continued activation of the proximal effectors leads to the up-regulation of a different collection of UPR-inducible proteins (Fig. 1D; Apoptosis), as well as the activation of other signaling pathways, that combine to promote cell death (Fig. 1C; JNK and caspase-12) [39, 40]. ER stress can also promote cell death in collaboration with the mitochondrial apoptosis pathway. For example, ER stress causes the release of cytochrome c [41]. Moreover, the pro-apoptotic Bcl-2 family members, Bax and Bak, associate with the ER, where they activate IRE1, thus linking mitochondrial and ER mediated apoptotic pathways [42]. Accordingly, the strength and duration of the ER stress contribute to determining the ultimate role of the UPR as either a survival or a death oriented signaling pathway.
One of the most thoroughly studied ER stress response genes is GRP78, which is also expressed in many cell types under non-stressed conditions, but upon activation of the UPR, is induced further. The GRP78 promoter has ER stress response elements (ERSEs) that bind XBP1 or ATF6, which is required for transcriptional induction during ER stress [14, 43, 44]. Like GRP78, most of the other ER stress response genes that have been characterized also have ERSEs [45], indicating that this transcriptional induction mechanism is highly conserved amongst genes that are induced during the UPR. Increased expression of GRP78 during the UPR enhances the protein folding capacity in the ER; if the load of misfolded proteins is reduced via this mechanism, by binding to the proximal effectors of ER stress, GRP78 contributes to inactivating the UPR, signaling resolution of the stress. Moreover, it was shown that during long-term ER stress, GRP78 redistributes from the ER to other locations, including the cytosol, where it can bind to and prevent the release of caspase-12 from the ER, thus inhibiting the apoptotic phase of the UPR [46].
Ischemia as an Activator of the UPR
Although studies employing chemicals to induce ER stress in cultured cells and, in a few cases, in vivo, have been useful in delineating the molecular details of the UPR, such conditions are relatively extreme, and are not likely to represent physiologically meaningful stresses. In contrast, the lack of oxygen and nutrients that take place during ischemia are known to affect the ER environment in ways predicted to activate the UPR. For example, glucose deprivation was one of the first maneuvers shown to activate the UPR, probably by impeding protein glycosylation in the ER, thus mimicking the effects of tunicamcyin [9, 10]. GRP78 was named a glucose-response protein based on its induction in cultured cells subjected to glucose starvation [47, 48]. Additionally, the machinery responsible for disulfide bond formation in the ER requires molecular oxygen for proper function [49].
Some of the earliest studies on the effects of ischemia/reperfusion on the UPR were carried out in the brain. For example, it was shown that in ischemic rabbit brain, several features of the UPR were activated, including PERK, eIF2□ phosphorylation, translational arrest and XBP1 mRNA splicing [50]. Moreover, ischemic pre-conditioning in the brain has been shown to induce GRP78 and to protect from further ischemic damage [51]. Gene array studies have shown that transient cerebral artery occlusion increases the expression of numerous UPR-dependent genes in the brain [52]. In addition to brain, numerous studies of the UPR have been carried out in tumors and cancer cell lines. For example, the UPR was shown to be activated in ischemic regions of tumors and in cultured tumor cells subjected to hypoxia; in both contexts, UPR activation was protective [53]. As a result of numerous studies in the tumor cell context, it has been suggested that the UPR provides a selective advantage to some aggressively growing solid tumors, where the rate of growth sometimes surpasses neo-angiogenesis, leading to ischemia and activation of the UPR [54]. In this context, since the UPR protects the growing tumor, and has been shown to contribute to malignant progression, the therapeutic strategy has been to inhibit the UPR in order to moderate tumor growth [55].
The demonstration that the UPR is activated in hypoxic tumor cells and tissue, as well as is the ischemic brain, prompted studies of whether the UPR is activated by ischemic in the heart, or by simulated ischemia in cultured cardiac myocytes. GRP78, was shown to increase in mouse hearts subjected to ex vivo ischemia/reperfusion, as well as in surviving cardiac myocytes that border the infarct zone in a mouse model of in vivo myocardial infarction [56]. Another study showed that GRP78 was induced and XBP1 was activated in hearts subjected to ischemia/reperfusion in vivo [57]. Transgenic overexpression of monocyte chemoattractant protein-1 (MCP-1) in mouse hearts was shown to induce ischemic heart disease and increase the expression of numerous ER stress response genes, including GRP78 [58]. Also, a number of ER stress response genes, including protein disulfide isomerase, were shown to be induced in the peri-infarct zone in a mouse model of myocardial infarction [59]. Several studies have shown that simulating ischemia or ischemia/reperufsion in cultured neonatal rat or adult mouse ventricular myocytes, or in the HL-1 atrial myocyte cell line, activates XBP1 and increases the expression of GRP78, and other genetic markers of the UPR [56, 57, 59-61].
Thus, ischemia and ischemia/reperfusion activate numerous features of the UPR in cardiac myocytes in vivo and in vitro; however, in contrast to studies in tumor cells and in the brain, where ER stress has been shown to be protective, it is less clear what function the UPR serves in the cardiac context. Some studies support protective roles for the UPR; for example, pre-activation of ATF6 in the hearts of transgenic mice was shown to protect the heart from ischemia/reperfusion damage [62], suggesting that under these conditions, genes induced by the ATF6 branch of the UPR served protective functions. Also, adenoviral-mediated overexpression of the ER stress response gene for protein disulfide isomerase decreased the size of infarcts in mouse hearts subjected to in vivo coronary artery ligation [59]. In other studies, it was shown that upregulation of GRP78 during ischemic pre-conditioning is responsible for protecting cultured cardiac myocytes from further ischemic injury [63], and that preinducing ER stress response genes with tunicamycin protects H9c2 cardiomyocytes from death induced by simulated ischemia/reperfusion [64]. It has also been shown that overexpression of the ER stress response gene, GRP94, protects cultured cardiac myocytes from death in response to simulated ischemia [65]. Taken together, these studies suggest that when the UPR is activated in the heart during ischemia, or ischemia/reperfusion, it may contribute to a protective stress response mounted by cardiac myocytes.
In contrast to the studies cited above, other results support the possibility that the UPR may contribute to ischemia/reperfusion damage in the heart. In cultured cardiac myocytes, it was shown that AMP kinase and ER stress were both activated during simulated ischemia, and that in this context, by inhibiting ER stress, AMP kinase protected the cells from hypoxic injury [60]. ER stress has been shown to activate autophagy-mediated cell death cardiac myocytes subjected to simulated ischemia/reperfusion [66]. Additionally, in cultured cardiac myocytes, ER stress leads to the activation of PKC delta, but that inhibiting delta PKC activation decreases ER stress-mediated apoptosis [57]. Overexpression of the ER stress response gene, p53-upregulated modulator of apoptosis (PUMA), increased apoptosis in cultured cardiomyocytes subjected to activation of the UPR [67], and targeted deletion of PUMA in mouse hearts was associated with reduced cardiomyoycte death upon ex vivo ischemia/reperfusion [68].
Although the reasons underlying the seemingly paradoxical results described above are not known, it is possible that like some other signaling pathways, the UPR can mediate both protective and damaging effects in the heart, depending upon the context. In support of this possibility is a study which showed that in cultured cardiac myocytes, simulated ischemia activated protective aspects of the UPR at early times, but at later times, apoptotic features of the UPR were dominant [61]. This finding is consistent with general views that during the initial phases of ER stress, the UPR mediates induction and activation of protective genes and proteins, but upon continued ER stress, pro-apoptotic machinery is activated. Although the mechanistic details of such a dual function for the UPR are still being worked out, one possibility is that ATF6, PERK and IRE-1 may be activated to different extents, depending upon the strength and nature of the ER stress, and that some effectors, e.g. ATF6, might mediate activation of mostly protective genes [33, 62], while others, e.g. PERK, may induce a greater number of pro-apoptotic genes. In a study designed to examine this possibility, it was shown that in cultured fibroblasts, even mild ER stress activates the 3 proximal effectors of the UPR to similar extents, which argues against selective activation of ER stress response effectors as a mechanism [69]. In the same study, it was shown that during mild ER stress, survival is favored due to the intrinsic instabilities of mRNAs and proteins that promote apoptosis compared to those that facilitate protein folding and adaptation. Thus, it is possible that in the heart, brief ischemic stress leads to a change in the UPR-regulated proteome that fosters protection, while more prolonged ischemia alters the proteome in ways that contribute to damage.
Future Directions
This review has focused on roles for the UPR in the ischemic myocardium, which are just beginning to be appreciated. In this context, many potentially important studies concerning the function of the UPR in the heart are yet to be carried out. For example, it will be important to determine what parameters dictate when the UPR fosters protection and when in contributes to damage in the ischemic heart. This information, coupled with a more complete understanding of the levels and functions of genes, and ultimately, UPR-regulated proteins, during ischemic stress, will be required in order to fully appreciate the impact of ER stress on myocardial function in the ischemic heart. It will also be important to determine how the UPR interacts with, and is influenced by the numerous other signaling pathways known to be activated in the ischemic heart, including hypoxia-inducible factor-1, AMP kinase, nitric oxide synthase, nuclear factor kappa B, as well as the mitogen activated protein kinase and protein kinase C families. Finally, there is evidence that in addition to ischemia, the UPR is activated in the heart under other conditions, including hypertrophy and heart failure [58, 70, 71]. Moreover, the UPR appears to be important for cardiac development [72, 73], as shown by the finding that the targeted deletion of the XBP1 in mice leads to embryonic lethality due to incomplete heart development [74]. Thus, it is apparent that the ER-associated UPR protein quality control system plays important roles in the normal heart, as well as the stressed and diseased heart, underscoring the importance of future studies aimed at elucidating the roles of this intricate signaling pathway in the heart.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Blobel G. Protein targeting. Biosci Rep. 2000 Oct;20(5):303–44. doi: 10.1023/a:1010318832604. [DOI] [PubMed] [Google Scholar]
- [2].Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006 Dec 8;99(12):1315–28. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- [3].Patterson C, Ike C, Willis PWt, Stouffer GA, Willis MS. The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation. 2007 Mar 20;115(11):1456–63. doi: 10.1161/CIRCULATIONAHA.106.649863. [DOI] [PubMed] [Google Scholar]
- [4].Drummond IA, Lee AS, Resendez E, Jr., Steinhardt RA. Depletion of intracellular calcium stores by calcium ionophore A23187 induces the genes for glucose-regulated proteins in hamster fibroblasts. J Biol Chem. 1987 Sep 15;262(26):12801–5. [PubMed] [Google Scholar]
- [5].Lee AS. Mammalian stress response: induction of the glucose-regulated protein family. Curr Opin Cell Biol. 1992 Apr;4(2):267–73. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- [6].Shamu CE, Cox JS, Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994 Feb;4(2):56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- [7].McMillan DR, Gething MJ, Sambrook J. The cellular response to unfolded proteins: intercompartmental signaling. Curr Opin Biotechnol. 1994 Oct;5(5):540–5. doi: 10.1016/0958-1669(94)90071-x. [DOI] [PubMed] [Google Scholar]
- [8].Kim YK, Lee AS. Transcriptional activation of the glucose-regulated protein genes and their heterologous fusion genes by beta-mercaptoethanol. Mol Cell Biol. 1987 Aug;7(8):2974–6. doi: 10.1128/mcb.7.8.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988 Mar 31;332(6163):462–4. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- [10].Dorner AJ, Wasley LC, Raney P, Haugejorden S, Green M, Kaufman RJ. The stress response in Chinese hamster ovary cells. Regulation of ERp72 and protein disulfide isomerase expression and secretion. J Biol Chem. 1990 Dec 15;265(35):22029–34. [PubMed] [Google Scholar]
- [11].Chang SC, Wooden SK, Nakaki T, Kim YK, Lin AY, Kung L, et al. Rat gene encoding the 78-kDa glucose-regulated protein GRP78: its regulatory sequences and the effect of protein glycosylation on its expression. Proc Natl Acad Sci U S A. 1987 Feb;84(3):680–4. doi: 10.1073/pnas.84.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998 Dec;18(12):7499–509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997 Sep;17(9):4957–66. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998 Dec 11;273(50):33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- [15].Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993 Jun 18;73(6):1197–206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- [16].Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993 Aug 27;74(4):743–56. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- [17].Kohno K. How Transmembrane Proteins Sense Endoplasmic Reticulum Stress. Antioxid Redox Signal. 2007 Sep 26; doi: 10.1089/ars.2007.1819. [DOI] [PubMed] [Google Scholar]
- [18].Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000 Jun;2(6):326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- [19].Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002 May 24;277(21):18728–35. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- [20].Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. Embo J. 1996 Jun 17;15(12):3028–39. [PMC free article] [PubMed] [Google Scholar]
- [21].Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997 Sep 19;90(6):1031–9. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- [22].Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005 Feb;25(3):921–32. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999 Jan 21;397(6716):271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- [24].Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002 Jan 3;415(6867):92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- [25].Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002 Jul;3(1):99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- [26].Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000 Dec;6(6):1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- [27].Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, et al. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001 Apr 1;355(Pt 1):19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, et al. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol. 2005 Feb;7(2):186–94. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- [29].Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997 Sep;17(9):5117–26. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].DenBoer LM, Hardy-Smith PW, Hogan MR, Cockram GP, Audas TE, Lu R. Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem Biophys Res Commun. 2005 May 27;331(1):113–9. doi: 10.1016/j.bbrc.2005.03.141. [DOI] [PubMed] [Google Scholar]
- [31].Stirling J, O’Hare P. CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol Biol Cell. 2006 Jan;17(1):413–26. doi: 10.1091/mbc.E05-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kondo S, Saito A, Hino S, Murakami T, Ogata M, Kanemoto S, et al. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol. 2007 Mar;27(5):1716–29. doi: 10.1128/MCB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007 Sep;13(3):351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- [34].Thuerauf DJ, Morrison L, Glembotski CC. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J Biol Chem. 2004 May 14;279(20):21078–84. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- [35].Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem. 2007 Aug 3;282(31):22865–78. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- [36].Brodsky JL. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem J. 2007 Jun 15;404(3):353–63. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005 Aug;7(8):766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- [38].Lord JM, Roberts LM, Stirling CJ. Quality control: another player joins the ERAD cast. Curr Biol. 2005 Dec 6;15(23):R963–4. doi: 10.1016/j.cub.2005.11.013. [DOI] [PubMed] [Google Scholar]
- [39].Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000 Jan 28;287(5453):664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- [40].Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, et al. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001 Apr 27;276(17):13935–40. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- [41].Hacki J, Egger L, Monney L, Conus S, Rosse T, Fellay I, et al. Apoptotic crosstalk between the endoplasmic reticulum and mitochondria controlled by Bcl-2. Oncogene. 2000 May 4;19(19):2286–95. doi: 10.1038/sj.onc.1203592. [DOI] [PubMed] [Google Scholar]
- [42].Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006 Apr 28;312(5773):572–6. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- [43].Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999 Nov;10(11):3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000 Sep 1;275(35):27013–20. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- [45].Thuerauf DJ, Hoover H, Meller J, Hernandez J, Su L, Andrews C, et al. Sarco/endoplasmic reticulum calcium ATPase-2 expression is regulated by ATF6 during the endoplasmic reticulum stress response: intracellular signaling of calcium stress in a cardiac myocyte model system. J Biol Chem. 2001 Dec 21;276(51):48309–17. doi: 10.1074/jbc.M107146200. [DOI] [PubMed] [Google Scholar]
- [46].Rao RV, Peel A, Logvinova A, del Rio G, Hermel E, Yokota T, et al. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002 Mar 13;514(23):122–8. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shiu RP, Pouyssegur J, Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteinsin Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3840–4. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Welch WJ, Garrels JI, Thomas GP, Lin JJ, Feramisco JR. Biochemical characterization of the mammalian stress proteins and identification of two stress proteins as glucose- and Ca2+-ionophore-regulated proteins. J Biol Chem. 1983 Jun 10;258(11):7102–11. [PubMed] [Google Scholar]
- [49].Gorlach A, Klappa P, Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal. 2006 Sep-Oct;8(910):1391–418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- [50].Paschen W. Endoplasmic reticulum dysfunction in brain pathology: critical role of protein synthesis. Curr Neurovasc Res. 2004 Apr;1(2):173–81. doi: 10.2174/1567202043480125. [DOI] [PubMed] [Google Scholar]
- [51].Hayashi T, Saito A, Okuno S, Ferrand-Drake M, Chan PH. Induction of GRP78 by ischemic preconditioning reduces endoplasmic reticulum stress and prevents delayed neuronal cell death. J Cereb Blood Flow Metab. 2003 Aug;23(8):949–61. doi: 10.1097/01.WCB.0000077641.41248.EA. [DOI] [PubMed] [Google Scholar]
- [52].Vikman P, Edvinsson L. Gene expression profiling in the human middle cerebral artery after cerebral ischemia. Eur J Neurol. 2006 Dec;13(12):1324–32. doi: 10.1111/j.1468-1331.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- [53].Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. Embo J. 2005 Oct 5;24(19):3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006 Feb;6(1):55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- [55].Feldman DE, Chauhan V, Koong AC. The unfolded protein response: a novel component of the hypoxic stress response in tumors. Mol Cancer Res. 2005 Nov;3(11):597–605. doi: 10.1158/1541-7786.MCR-05-0221. [DOI] [PubMed] [Google Scholar]
- [56].Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res. 2006 Aug 4;99(3):275–82. doi: 10.1161/01.RES.0000233317.70421.03. [DOI] [PubMed] [Google Scholar]
- [57].Qi X, Vallentin A, Churchill E, Mochly-Rosen D. deltaPKC participates in the endoplasmic reticulum stress-induced response in cultured cardiac myocytes and ischemic heart. J Mol Cell Cardiol. 2007 Oct;43(4):420–8. doi: 10.1016/j.yjmcc.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol. 2006 Sep;291(3):H1411–20. doi: 10.1152/ajpheart.01378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Severino A, Campioni M, Straino S, Salloum FN, Schmidt N, Herbrand U, et al. Identification of protein disulfide isomerase as a cardiomyocyte survival factor in ischemic cardiomyopathy. J Am Coll Cardiol. 2007 Sep 11;50(11):1029–37. doi: 10.1016/j.jacc.2007.06.006. [DOI] [PubMed] [Google Scholar]
- [60].Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, et al. AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol. 2005 Nov;25(21):9554–75. doi: 10.1128/MCB.25.21.9554-9575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Szegezdi E, Duffy A, O’Mahoney ME, Logue SE, Mylotte LA, O’Brien T, et al. ER stress contributes to ischemia-induced cardiomyocyte apoptosis. Biochem Biophys Res Commun. 2006 Nov 3;349(4):1406–11. doi: 10.1016/j.bbrc.2006.09.009. [DOI] [PubMed] [Google Scholar]
- [62].Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, et al. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006 May 12;98(9):1186–93. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- [63].Shintani-Ishida K, Nakajima M, Uemura K, Yoshida K. Ischemic preconditioning protects cardiomyocytes against ischemic injury by inducing GRP78. Biochem Biophys Res Commun. 2006 Jul 14;345(4):1600–5. doi: 10.1016/j.bbrc.2006.05.077. [DOI] [PubMed] [Google Scholar]
- [64].Zhang PL, Lun M, Teng J, Huang J, Blasick TM, Yin L, et al. Preinduced molecular chaperones in the endoplasmic reticulum protect cardiomyocytes from lethal injury. Ann Clin Lab Sci. 2004 Autumn;34(4):449–57. [PubMed] [Google Scholar]
- [65].Vitadello M, Penzo D, Petronilli V, Michieli G, Gomirato S, Menabo R, et al. Overexpression of the stress protein Grp94 reduces cardiomyocyte necrosis due to calcium overload and simulated ischemia. Faseb J. 2003 May;17(8):923–5. doi: 10.1096/fj.02-0644fje. [DOI] [PubMed] [Google Scholar]
- [66].Takagi H, Matsui Y, Sadoshima J. The role of autophagy in mediating cell survival and death during ischemia and reperfusion in the heart. Antioxid Redox Signal. 2007 Sep;9(9):1373–81. doi: 10.1089/ars.2007.1689. [DOI] [PubMed] [Google Scholar]
- [67].Nickson P, Toth A, Erhardt P. PUMA is critical for neonatal cardiomyocyte apoptosis induced by endoplasmic reticulum stress. Cardiovasc Res. 2007 Jan 1;73(1):48–56. doi: 10.1016/j.cardiores.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Toth A, Jeffers JR, Nickson P, Min JY, Morgan JP, Zambetti GP, et al. Targeted deletion of Puma attenuates cardiomyocyte death and improves cardiac function during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2006 Jul;291(1):H52–60. doi: 10.1152/ajpheart.01046.2005. [DOI] [PubMed] [Google Scholar]
- [69].Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, et al. Adaptation to ER stress is mediated by differential stabilities of prosurvival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006 Nov;4(11):e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, et al. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004 Aug 10;110(6):705–12. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- [71].Hamada H, Suzuki M, Yuasa S, Mimura N, Shinozuka N, Takada Y, et al. Dilated cardiomyopathy caused by aberrant endoplasmic reticulum quality control in mutant KDEL receptor transgenic mice. Mol Cell Biol. 2004 Sep;24(18):8007–17. doi: 10.1128/MCB.24.18.8007-8017.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Barnes JA, Smoak IW. Glucose-regulated protein 78 (GRP78) is elevated in embryonic mouse heart and induced following hypoglycemic stress. Anat Embryol (Berl) 2000 Jul;202(1):67–74. doi: 10.1007/s004290000090. [DOI] [PubMed] [Google Scholar]
- [73].Mao C, Tai WC, Bai Y, Poizat C, Lee AS. In vivo regulation of Grp78/BiP transcription in the embryonic heart: role of the endoplasmic reticulum stress response element and GATA-4. J Biol Chem. 2006 Mar 31;281(13):8877–87. doi: 10.1074/jbc.M505784200. [DOI] [PubMed] [Google Scholar]
- [74].Masaki T, Yoshida M, Noguchi S. Targeted disruption of CRE-binding factor TREB5 gene leads to cellular necrosis in cardiac myocytes at the embryonic stage. Biochem Biophys Res Commun. 1999 Aug 2;261(2):350–6. doi: 10.1006/bbrc.1999.0972. [DOI] [PubMed] [Google Scholar]