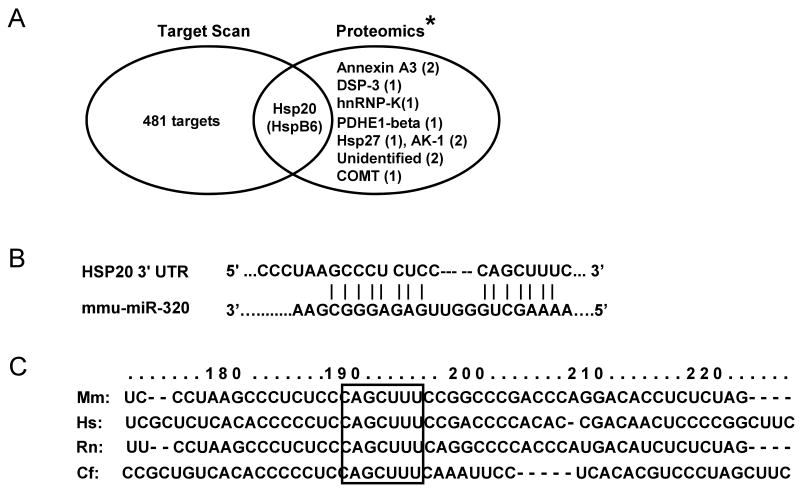
Figure 5.
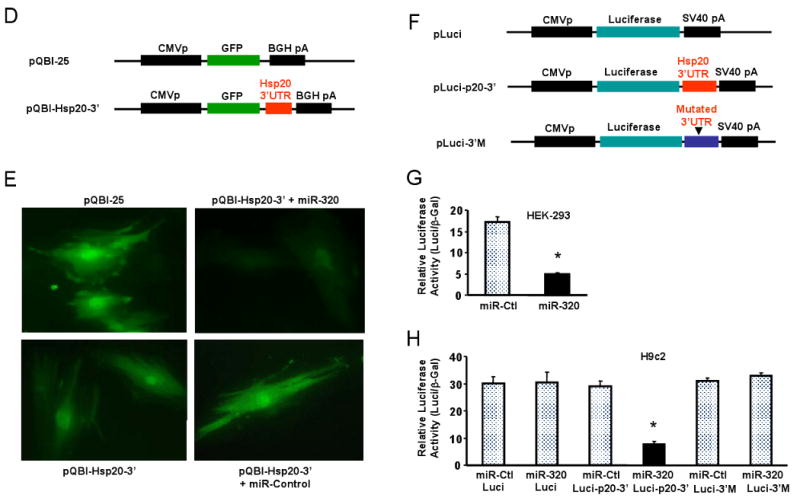
Hsp20 is a target of miR-320 in the murine heart. (A) TargetScan predicts that miR-320 has 482 targets, however, proteomic data showed that only 12 protein spots were altered in the proteome of I/R hearts (* Ref.# 22), and Hsp20 was listed in both the TargetScan results and the proteomics analysis. (B) Sequence alignment of miR-320 and 3′UTR of Hsp20. Note the complementarity at the 5′ and 3′ end of miR-320, where the crucial seed regions are located. (C) The putative miR-320-binding sites within the Hsp20 3′UTR are conserved among mammalian species (mouse, human, rat and dog). (D) Diagram of plasmid construction. A segment of Hsp20 3′ UTR was inserted downstream of the GFP-encoding sequence. (E) H9c2 cells were co-transfected with the plasmid containing the segment of Hsp20 3′ UTR and either miR-320 or a control oligoribonucleotide. Pictures were taken 48 h after transfection. (F) Diagram of plasmid construction. A segment of Hsp20 3′ UTR or a mutated segment was cloned downstream of the luciferase-encoding region. (G) Dual luciferase activity assay of HEK-293 cells co-transfected with the plasmid containing the segment of Hsp20 3′ UTR and either miR-320 or a control oligoribonucleotide showed that miR-320 inhibited luciferase activity, compared with controls. (H) Luciferase activity in H9c2 cells co-transfected with the various vectors indicated. (* P<0.05 relative to respective controls). Similar results were observed in three additional, independent experiments.