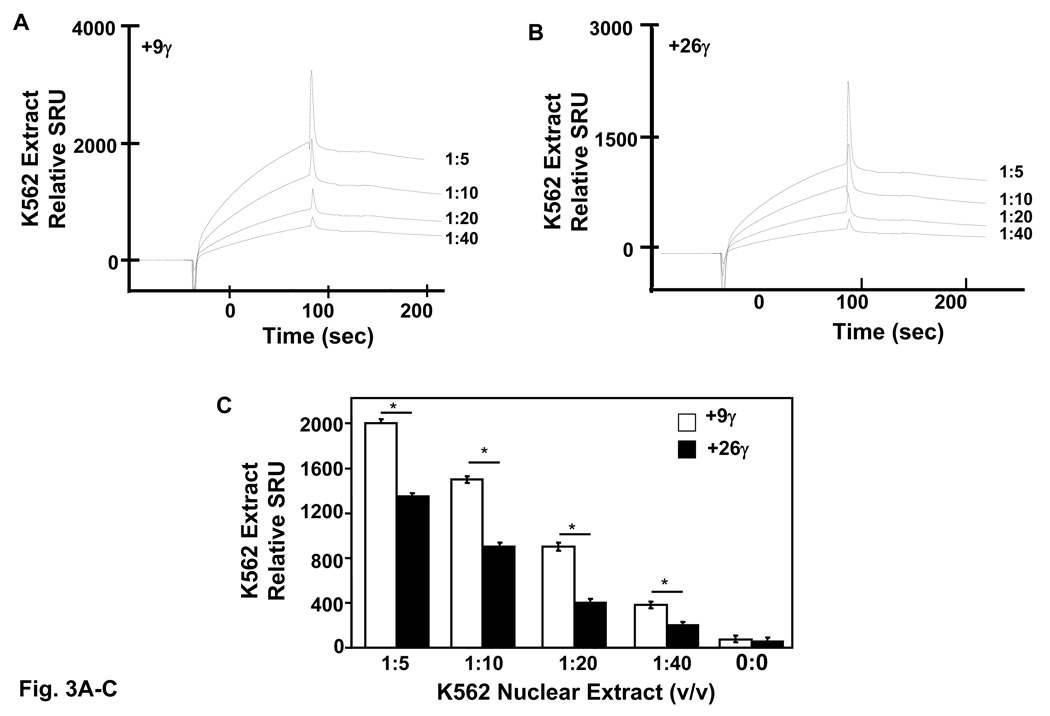
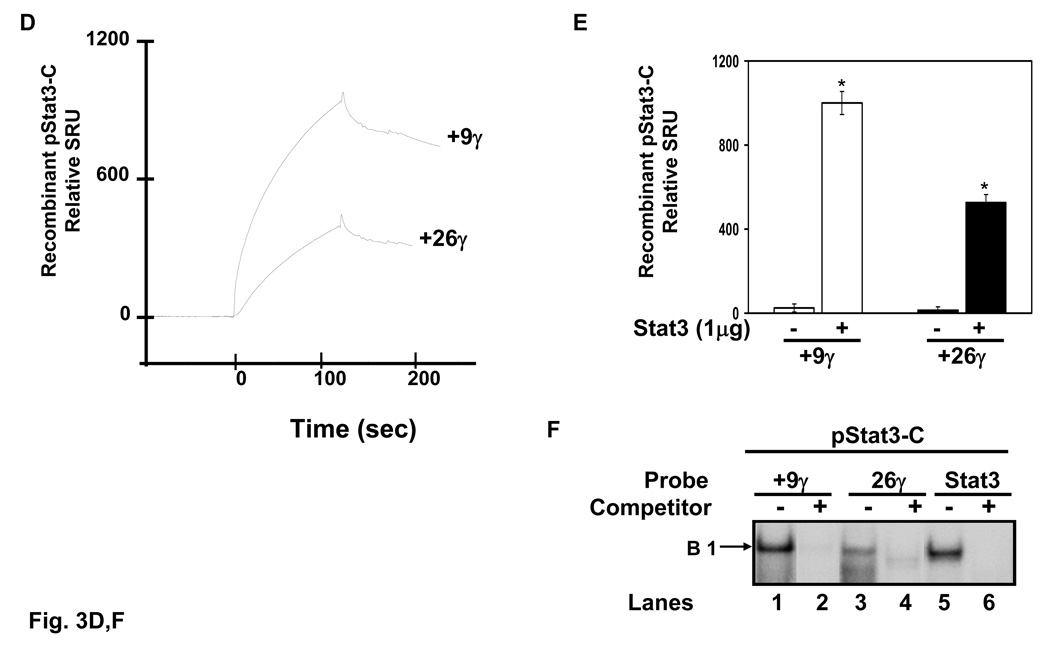
Fig. 3. High and low-affinity Stat3 binding sites exists in the γ-globin 5’UTR.
Quantitative assessment of DNA-protein interactions was performed with two probes, +9γ (+11 to +18) and +26γ (+23 to +37) by SPR analysis (see materials and methods). Biotin-conjugated probes were separately bound to streptavidin-coated sensor chips and treated with K562 nuclear extract or recombinant activated Stat3 protein. The level of protein binding to target consensus sequence was measured in SPR response units (SRU). A) The +9γ probe was analyzed with 1:5 to 1:40 dilutions of K562 nuclear proteins. Note the concentration-dependent increase in RU readout. B) The same studies described in panel A were completed with the +26γ probe. C) Shown in the graph is the quantification for protein bound to the γ-globin probes from panels A and B. Binding studies were performed without nuclear extract (0:0) as a control. The results are expressed as the mean ± SEM. The line above the graphs indicates a significant difference in binding between the two probes at p<0.05. D) A graph showing the relative RU generated by SPR analysis with activated pStat3 (pStat3-C) protein (1 µg). E) Quantitative data collected from the SPR analysis completed in panel D. Binding studies were performed in the presence and absence of recombinant pStat3-C protein. F) EMSA using recombinant Stat3 protein and the γ-globin probes showed the formation of a single DNA-protein complex with +9γ and consensus Stat3 probes in lanes 1 and 5 respectively. Formation of this complex was abolished by cold self oligo competitor (lanes 2 and 6). Similar studies were performed with the +26γ probe which produced a less prominent binding pattern which was competed by cold self oligo (lanes 3 and 4).