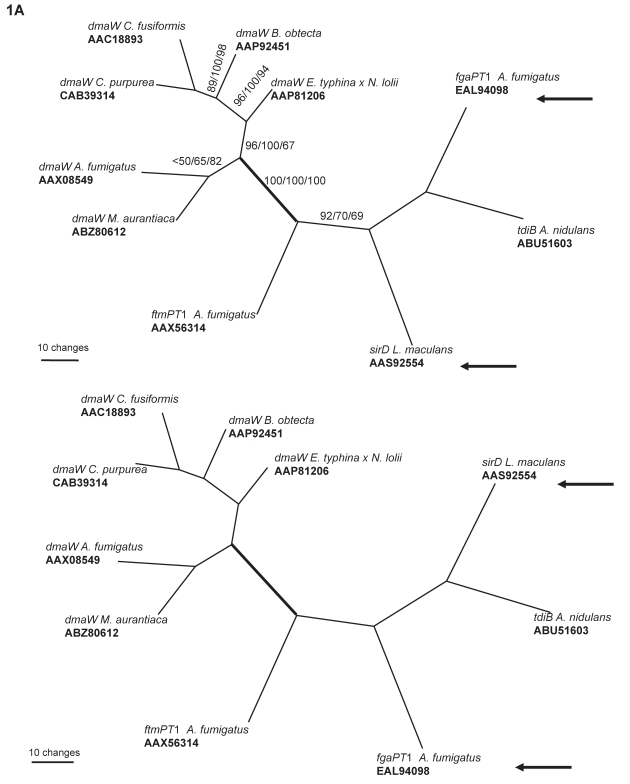
Figure 1.
The most parsimonious trees relating amino acid sequences of the deduced products of six dmaW genes and four related genes with known functions. A) Tree based on whole gene region. Of 653 aligned characters, 250 characters are informative. Length = 1521, CI = 0.870, RI = 0.503. Arrows show the discrepancies of the two most parsimonious trees. B and C) Trees based on relatively conserved gene regions screened by GBlocks. Of 308 total aligned characters, 166 characters are informative. Length = 934, CI = 0.864, RI = 0.512. Products of dmaW genes formed a monophyletic group with 100% bootstrap support. B. Unrooted tree; C. Phylogram rooted by choosing the non-dmaW gene products as the outgroup. numbers on branches indicate bootstrap percentage of MP/posterior probabilities of BI/bootstrap percentage of ML; *indicates that particular branch does not exist in the analysis. The thick branch separates of the outgroup from the ingroup.