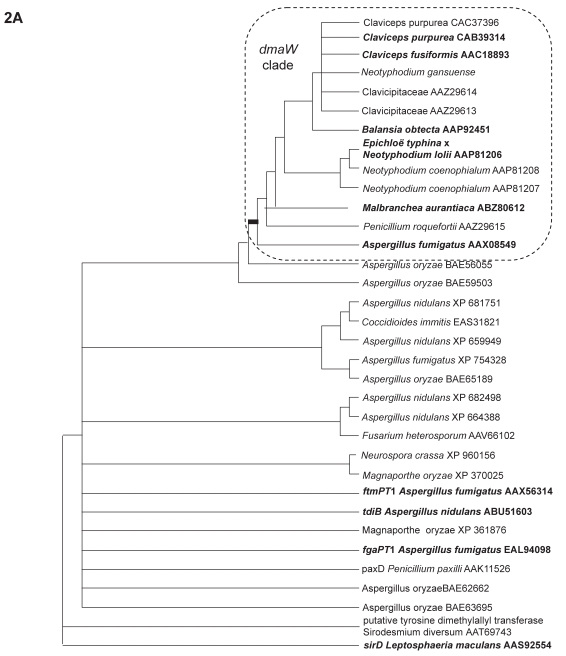
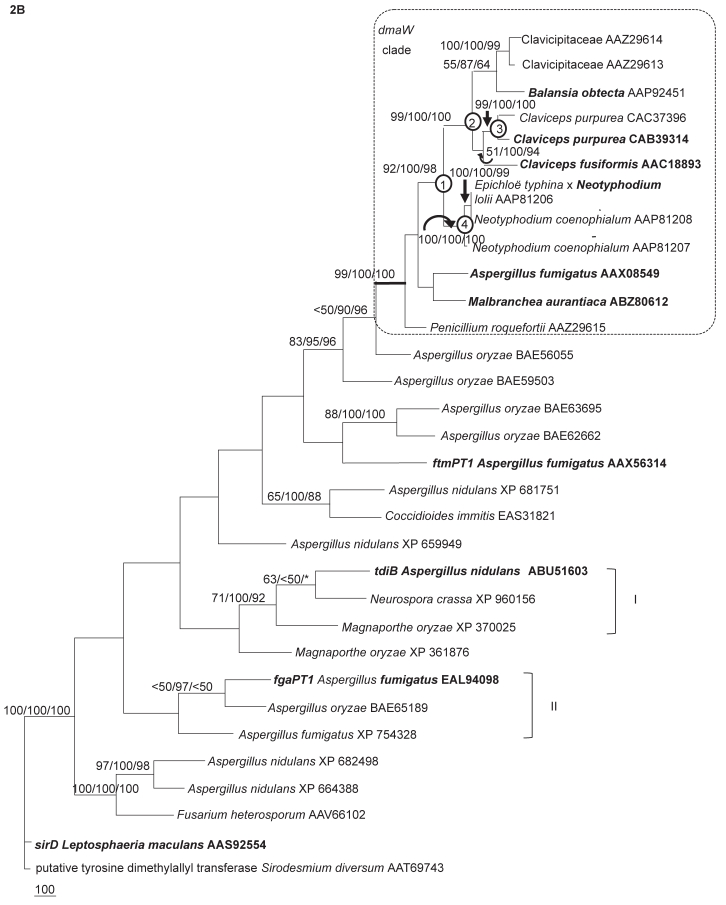
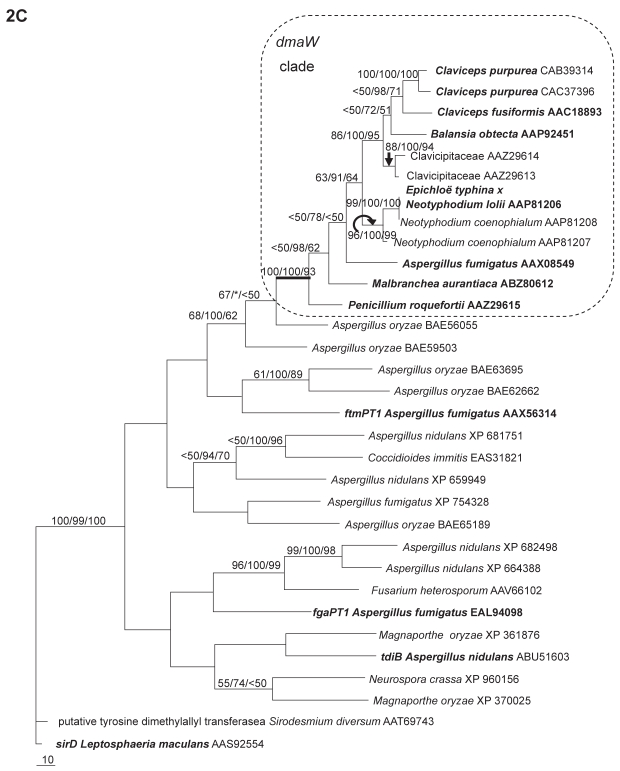
Figure 2.
A) Strict consensus tree of the six most parsimonious trees for dmaW gene products and products of multiple related genes of 34 OTUs based on relatively conserved gene regions. Of the 155 characters, 133 were parsimony-informative; length = 1366, CI = 0.594, RI = 0.499. B) One of the two most parsimonious trees of 32 OTUs based on the whole gene region. Of the 933 characters, 456 were parsimony-informative characters. Length = 4974, CI = 0.682, RI = 0.499. C) The most parsimonious trees for dmaW gene products and products of multiple related genes of 32 OTUs based on relatively conserved gene regions, which were screened by Gblocks. Total aligned characters = 155, informative characters = 135; length = 1427, CI = 0.607, RI = 0.497. OTUs in bold indicate the gene products with functions that have been confirmed. Numbers on branches indicate bootstrap percentage of MP/posterior probabilities of BI/bootstrap percentage of ML; *indicates that particular branch does not exist in the analysis; numbers in circles indicate the possible gene duplication events (also see Fig. 3).