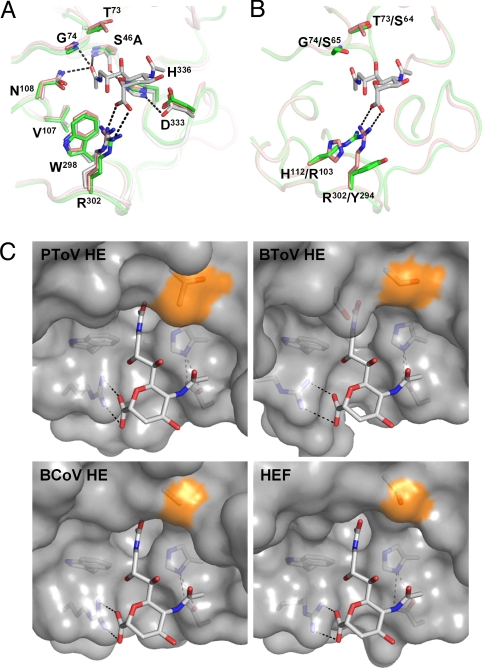
Fig. 3.
Structure of the PToV and BToV HE esterase sites and comparison to BCoV HE and HEF. (A) Superposition of PToV HE (orange), BCoV HE (green) and HEF (gray). Substrate analog 9-N-acetyl sialic acid (Neu5,9NAc2α2Me) present in the structure of HEF is shown in stick representation (carbon, gray; nitrogen, blue; oxygen, red). Hydrogen bonds in the catalytic triad and oxyanion hole of HEF are indicated by dashed black lines. Residue labeling refers to PToV HE0 that carries an active-site Ser46Ala mutation. (B) Superposition of the active sites of BToV HE (green) and PToV HE (orange). As shown, swapping of the active-site arginine between positions 294 and 103 in BToV HE would preserve hydrogen bonding to the sialoside carboxylate; note that optimal bidentate hydrogen bonding between Arg103 and the Sia carboxylate would require a ≈50° rotation and a 1.5-Å shift of the substrate's pyranose ring. Hydrogen bonds are indicated as in B. Residue labeling refers to PToV HE/BToV HE. (C) Surface representation of the PToV HE, BToV HE, BCoV HE, and HEF active sites with surface patches contributed by Thr73, Ser64, Ala74, and Ser84 in the respective proteins colored in orange. Note that the presence of Thr in PToV HE reduces the amount of free space available for substituents at the Sia C7 position, which, as shown by enzymatic analysis (Fig. 4), fully determines PToV HE substrate preference for 9-mono-O-acetylated Sias.