Figure 1. The structure and function of chimeric SRK genes.
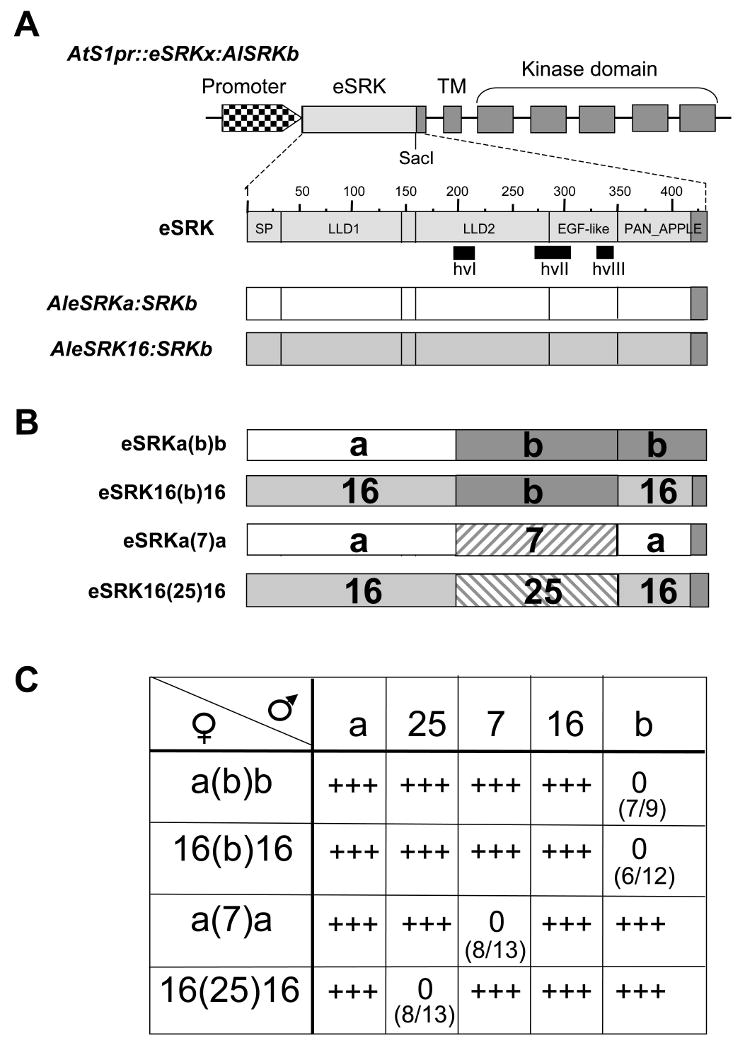
A. The AtS1pr∷eSRKx:AlSRKb genes used for construction of eSRK chimeras. The top diagram shows the structure of a generic AtS1pr∷eSRKx:AlSRKb gene, in which the AtS1 promoter (checkered arrowhead) drives an SRK transcriptional unit with its seven exons and native 3′ untranslated sequences. Exon 1 encodes the AlSRK extracellular domain (eSRK), exon 2 encodes the transmembrane domain (TM), and exons 3-7 encode the kinase domain. The unique SacI restriction site used for construction of chimeras is shown towards the 3′ end of the eSRK. AlSRKb sequences are shown in dark grey and eSRK sequences (from the initiating methionine codon to the unique SacI site) derived from other variants are shown in light grey. Due to the use of the SacI site, all constructs used in this study contain a common 23-amino-acid region derived from AlSRKb (shown in dark grey, spanning the last 23 amino acids of eSRK, i.e. residues 411-434 in SRKb). The middle diagram is a magnified view of the AleSRKb (with numbers indicating amino acids). The vertical lines delineate predicted structural subdomains in the eSRK [29]: SP, signal peptide; LLD1 and LLD2, lectin-like domains 1 and 2; EGF-like, epidermal growth factor-like domain; and PAN_APPLE domain. The locations of hypervariable regions are indicated below the diagrams and correspond to the following amino-acid segments in AlSRKb: 204-219 (hvI), 269-304 (hvII), 326-340 (hvIII). The lower diagrams show the eSRKs of AtS1pr∷AleSRKa:AlSRKb and AtS1pr∷AleSRK16:AlSRKb, two of the constructs used for domain swaps.
B. eSRK chimeras. The structures of four functional chimeric eSRKs are shown. The derivation of various segments is shown by different colors or patterns: AleSRKb: grey; AleSRKa: white; AleSRK16: hatched; AleSRK25: stippled; CgeSRK7: slanted bricks. The limits of the swapped region in these and other chimeras analyzed are indicated in Table 1 and their sequences are shown in Figure S2.
C. Pollination phenotypes of A. thaliana plants transformed with eSRK chimeras. First- and second-generation transgenic plants expressing each chimera were pollinated using plants expressing the cognate SCR, other SCRs, and wild-type pollen. eSRK chimeras are indicated in the column below the female symbol: a(b)b, AleSRKa(b)b:AlSRKb; 16(b)16, AleSRK16(b)16:AlSRKb; a(7)a, CgeSRKa(7)a:AlSRKb; 16(25)16, AleSRK16(25)16:AlSRKb. The SCR variants expressed in pollen used for pollination assays are indicated in the row to the right of the male symbol and correspond to the constructs shown in Figure S1: a, native AlSCRa; 25, native AlSCR25; 7, AlSCRb:CgSCR7; 16, AlSCRb:AlSCR16; b, native AlSCRb. The numbers in parentheses show the number of T1 plants that expressed an incompatibility response towards pollen expressing cognate SCR over the total number of primary transformants analyzed. 0 = an incompatible response (typically <5 pollen tubes per pollinated stigma); +++ = a compatible response (typically >50 pollen tubes per pollinated stigma). For each construct, although the majority of transformants exhibiting SI expressed a strong SI response (<5 pollen tubes per pollinated stigma), typically 1 or 2 transformants exhibited a weaker SI response (5-10 pollen tubes per pollinated stigma).
