Figure 2. Structure and immunoblot analysis of eSRKa(7)a:SRKb and eSRK16(25)16:SRKb substitution mutants.
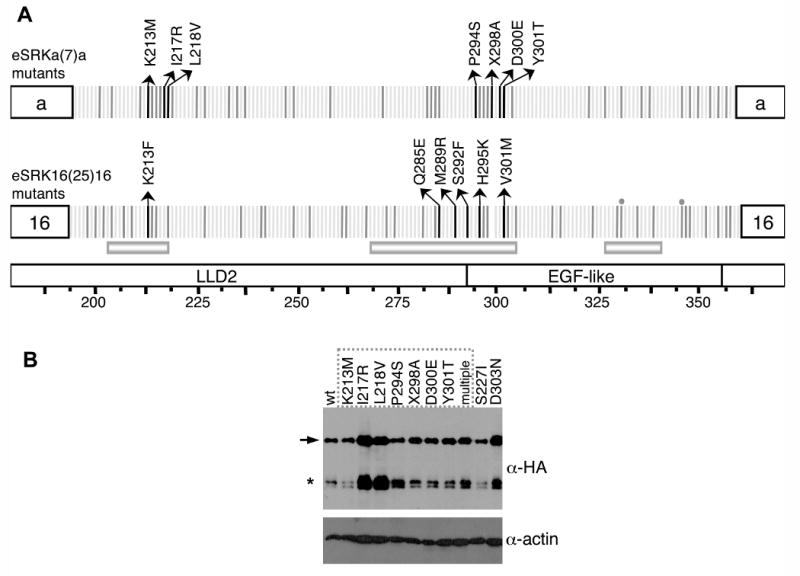
A. Single-codon substitutions in eSRK chimeras. The diagrams show the specificity-determining hvI-hvIII regions of the eSRKa(7)a and eSRK16(25)16 chimeras, with amino-acid residues depicted by vertical bars. The bottom diagram shows the location of the LLD2 and EGF-like domains and hypervariable regions hvI, hvII, and hvIII. Residues that do not differ between eSRKa and eSRKa(7)a or between eSRK16 and eSRK16(25)16 are shown by light grey bars. Polymorphic residues that were modified by substitution mutagenesis are shown by dark grey and black bars. Each of these variable residues, with the exception of 2 residues in eSRK16(25)16 (marked by gray circles, mutants of which failed to generate transgenic plants), were individually replaced in eSRKa(7)a with residues found at the equivalent positions in eSRKa, and in eSRK16(25)16 with residues found at the equivalent positions in eSRK16. Transgenic stigmas expressing each of the single-codon substitution eSRKa(7)a or eSRK16(25)16 derivatives were tested by pollination with SCR7- or SCR15-expressing pollen, respectively. For most substitution mutants (substituted residues shown as dark grey bars), the stigmas of at least some transformants [13-88% of transformants for eSRKa(7)a mutants and 12-71% of transformants for eSRK16(25)16 mutants] exhibited an incompatible response. For each of the substitution mutants that failed to confer an incompatibility response, between 10 and 18 independent transformants were assayed. Amino-acid residues found to be required for the function of eSRK chimeras are shown by black bars with arrows indicating the amino-acid substitution that caused loss of chimera function. The L218V substitution in eSRKa(7)a and the Q285E substitution in eSRK16(25)16 conferred a weakened incompatibility response (Figure S3). The X298A change in eSRKa(7)a involved inserting alanine at amino-acid site 298. Note that substitutions at two sites, 213 and 301, disrupted the function of both eSRKa(7) and eSRK16(25)16 chimeras: site 213 was sensitive to a change from the polar and charged lysine to a non-polar and uncharged methionine or phenylalanine, while site 301 was sensitive to changes in volume.
B. Immunoblot analysis of eSRK chimeras. For immunoblot analysis of HA-tagged eSRKa(7)a chimeras, proteins were extracted from the stigmas of plants transformed with AtS1pr∷eSRKa(7)a:AlSRKb (wt) and its single- and multiple-codon substitution derivatives, and subjected to protein immunoblot analysis (Supplemental Data). The “wt” lane shows the level of non-mutated “wild type” eSRKa(7)a protein found in stigmas exhibiting an incompatibility response toward SCR7-expressing pollen. The remaining lanes show representative patterns for eSRKa(7)a substitution derivatives: nine single-codon substitution derivatives labeled according to the amino-acid substitution introduced into each chimera (numbering as in panel A), and a multiple-codon substitution derivative (multiple). The dashed box indicates the substitution derivatives that did not confer an incompatible response towards SCR7-expressing pollen. The blot was probed sequentially with an anti-HA monoclonal antibody (top panel) and an anti-actin antibody as loading control (bottom panel). The arrow indicates the full-length eSRKa(7)a:SRKb receptor and the asterisk indicates the alternative smaller SRK products typically produced in stigmas [39].
