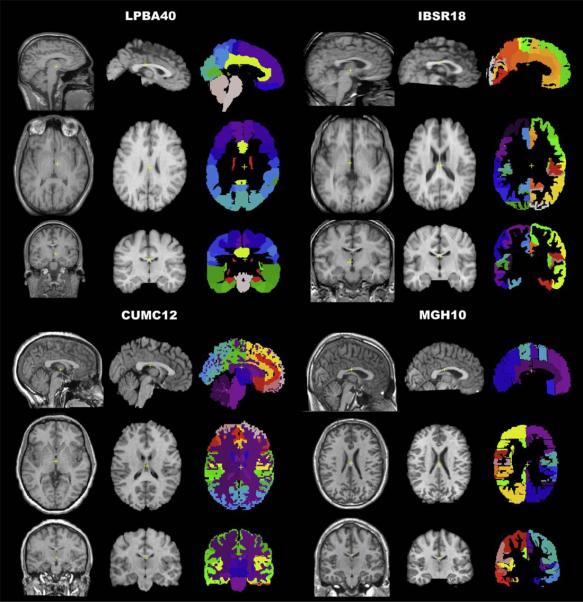
Fig. 1.
Brain image data. The study used four different image datasets with a total of 80 brains. The datasets contain different numbers of subjects (n) and different numbers of labeled anatomical regions (r) derived from different labeling protocols: LPBA40 (LONI Probabilistic Brain Atlas: n=40, r=56), IBSR18 (Internet Brain Segmentation Repository: n=18, r=84), CUMC12 (Columbia University Medical Center: n=12, r=128), and MGH10 (Massachusetts General Hospital: n=10, r=74). A sample brain from each dataset is shown. For each brain, there are three columns (left to right): original T1-weighted MRI, extracted brain registered to nonlinear MNI152 space, and manual labels registered to nonlinear MNI152 space (used to extract the brain). Within each column the three rows (top to bottom) correspond to sagittal (front facing right), horizontal (front facing top, right on right side), and coronal (right on right side) views. The LPBA40 brains had already been extracted and registered to MNI (MNI305 vs. MNI152) space (Shattuck et al., 2008). The scale, position, and contrast of the MR images have been altered for the figure. The colors for the manual labels do not correspond across datasets. (In the Discussion the reader is referred to the web version of this article.)