Figure 2.
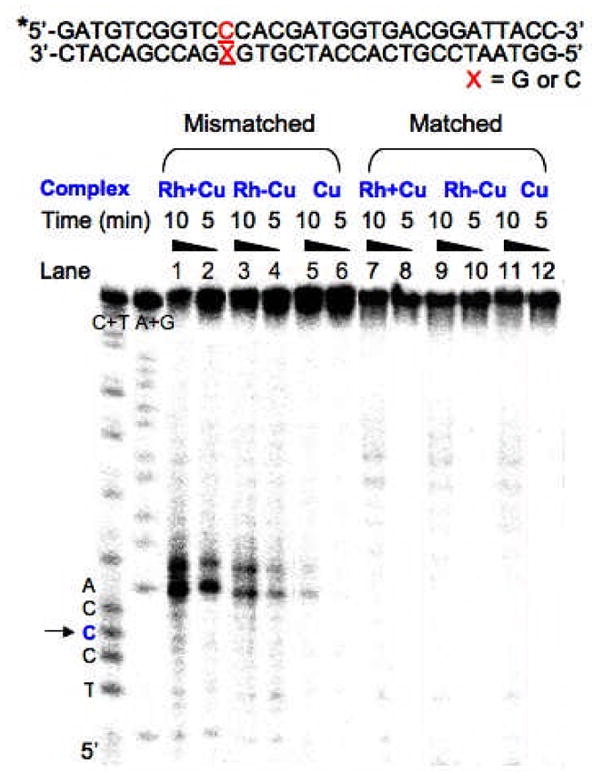
Sequences of 31mer oligonucleotides (top). Autoradiogram of a denaturing 20% polyacrylamide gel presenting DNA cleavage by Cu, Rh+Cu and Rh−Cu with matched and mismatched oligonucleotides (bottom). A+G and C+T, Maxam-Gilbert sequencing reactions. * indicates 5′-32P-end-label of the strand. Conditions: duplex (1 μM), Cu (1 μM CuCl2, 3 μM phen, 5 mM sodium ascorbate), Rh+Cu (1 μM [Rh(bpy)2(chrysi)]3+, 1 μM CuCl2, 3 μM phen, 5 mM sodium ascorbate), Rh−Cu (1 μM [Rh(chrysi)(phen)(bphen)]3+, 1 μM CuCl2, 3 μM phen, 5 mM sodium ascorbate), in 10 mM Tris, pH 7.5, 50 mM NaCl at 37 oC. Lanes 1–2 and 7–8, fragments with Rh+Cu; lanes 3–4 and 9–10, fragments with Rh−Cu; lanes 5–6 and 11–12, fragment with Cu. The time shown reflects incubation time after treatment with sodium ascorbate before quenching. The arrow marks the mismatched site.
