Figure 3.
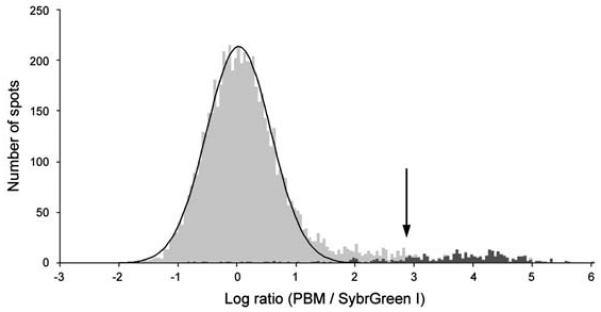
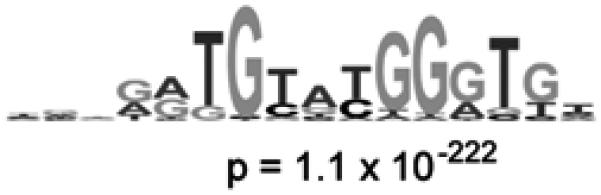
Identification of the DNA binding site motif from the significantly bound spots. (a) Distribution of ratios of PBM data, normalized by SYBR Green I data, for the yeast TF Rap1 bound to yeast intergenic microarrays. The arrow indicates those spots passing a P-value cutoff of 0.001 after correction for multiple hypothesis testing. Indicated in dark gray are spots with an exact match to a sequence belonging to the PBM-derived binding site motif. (b) Sequence logo (Schneider and Stephens, 1990) of the PBM-derived motif for the yeast transcription factor Rap1. (Reproduced from (Mukherjee et al., 2004) with permission from Nature Publishing Group.)
