Figure 3.
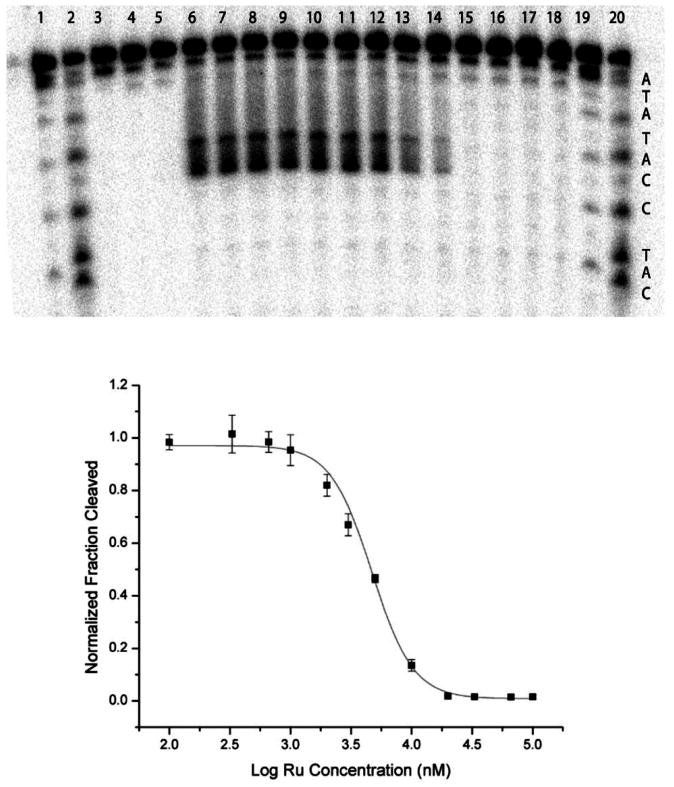
Competitive binding of Ru(bpy)2(eilatin)2+ to mismatched DNA monitored using Rh(bpy)2(chrysi)3+ photocleavage. Top: Denaturing polyacrylamide gel of a competition experiment between Rh(bpy)2(chrysi)3+ and Ru(bpy)2(eilatin)2+ for a CC mismatch in the oligonucleotide 5′-32P-TTAGGATCATCCATATA-3′. AG and CT lanes are Maxam–Gilbert sequencing reactions. All samples contained a 3 μM mismatched duplex in a buffer of 50 mM NaCl, 10 mM NaPi, and pH 7.1 and were irradiated for 10 min using a solar simulator unless otherwise stated. Sample conditions: lane 1, DNA only irradiated without Rh; lane 2, 3 μM Rh(bpy)2(chrysi)3+ without irradiation; lane 3, 3 μM Ru(bpy)2(eilatin)2+ irradiated without Rh; lanes 4–16, 3 μM Rh(bpy)2(chrysi)3+ and increasing concentrations of Ru(bpy)2(eilatin)2+, 0, 0.1, 0.33, 0.66, 1, 2, 3, 5, 10, 20, 33, 66, and 100 μM, respectively. Wide photocleavage bands do not reflect nonspecific photocleavage at more than one site but rather the multiple products produced by hydrogen abstraction upon photoactivated cleavage at the mismatched site. Bottom: Plot of a fraction cleaved against the Ru(bpy)2(eilatin)2+ concentration for four trials of the competition experiment.
