Figure 2.
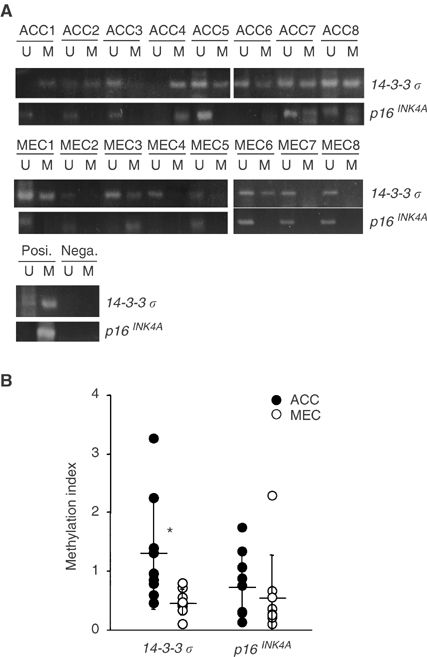
Methylation-specific PCR of the CpG island of the 14-3-3 σ and p16INK4A genes in ACC and MEC. (A) Microdissected tissues were subjected to MSP, and electrophoresis was performed in 4% agarose gels. The human fibrosarcoma cell line HT1080 was used as a positive control for methylated 14-3-3 σ. Normal salivary gland cells (SG) treated with SssI methyltransferase were used as a positive control for p16INK4A. Water was used as a negative control. (B) Methylation of the 14-3-3 σ and p16INK4A genes was semiquantitated by densitometric analysis. Methylation indices were calculated by dividing the density of the unmethylated allele into that of the methylated allele. Horizontal bars and vertical bars show mean and s.d., respectively. *Statistically significant by one-way ANOVA (*P=0.0304).
