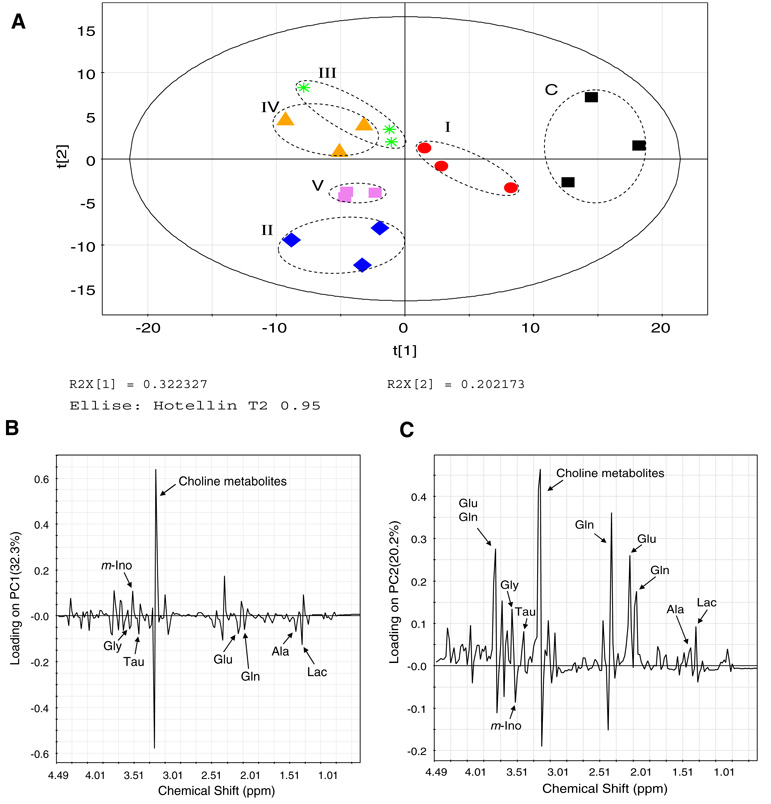
Fig. 2.
Principal component analysis of the metabolomes following all six treatments. (a) A PCA score plot distinguishes the metabolic profiles of SH-SY5Y neuroblastoma cells treated with: I: 1 mM MPTP; II: 5 mM 3-NP; III: 4 µg/ml antimycin-A; IV: 2 mM sodium azide and V: 1 µg/ml oligomycin. C: Control cells treated with saline only. The solid ellipse denotes the 95% significance limit (Hotelling T2) and the dotted ellipse indicates the differentially inhibited sample groups (the dotted lines do not have statistical significance). (b) Loading plot for principal component 1 (PC1). (c) Loading plot for principal component 2 (PC2). The PC1 calculated by this method represents the largest variation among the data sets and the subsequent PCs represent lesser variations by comparison. Usually, the first three PCs should be able to explain 50–80% of the total variation. The resulting PCA scores were subjected to one-way analysis of variance (ANOVA) in order to test the level of statistical difference among the sample groups. Differences were considered significant when P ≤ 0.05