Abstract
The specificity of vesicle-mediated transport is largely regulated by the membrane-specific distribution of SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins. However, the signals and machineries involved in SNARE protein targeting to the respective intracellular locations are not fully understood. We have identified a Sec22 ortholog in Plasmodium falciparum (PfSec22) that contains an atypical insertion of the Plasmodium export element within the N-terminal longin domain. This Sec22 protein partially associates with membrane structures in the parasitized erythrocytes when expressed under the control of the endogenous promoter element. Our studies indicate that the atypical longin domain contains signals that are required for both endoplasmic reticulum (ER)/Golgi apparatus recycling of PfSec22 and partial export beyond the ER/Golgi apparatus interface. ER exit of PfSec22 is regulated by motifs within the α3 segment of the longin domain, whereas the recycling and export signals require residues within the N-terminal hydrophobic segment. Our data suggest that the longin domain of PfSec22 exhibits major differences from the yeast and mammalian orthologs, perhaps indicative of a novel mechanism for Sec22 trafficking in malaria parasites.
Plasmodium falciparum exhibits a complex network of endomembrane organelles that are unique to this obligate intracellular parasite of human erythrocytes. They include parasite-induced tubules and vesicles in the infected host cell and specialized secretory structures collectively known as the apical complex. The asexual blood stages of the parasite develop within a parasitophorous vacuole (PV) and thus are separated from the external milieu by three lipid bilayers: the parasite plasma membrane (PPM), the PV membrane (PVM), and the erythrocyte plasma membrane. To survive inside these terminally differentiated human erythrocytes, P. falciparum remodels the host cell compartment by exporting numerous proteins into the erythrocyte cytoplasm (12, 15, 49, 50, 57). The mechanisms by which both soluble and membrane-bound proteins are transported, first into the PV lumen, followed by translocation across the PVM and transport within the erythrocyte cytosol, are not fully understood (9). A majority of the exported proteins contain bipartite signals that comprise a “recessed” N-terminal signal sequence and a Plasmodium export element/vacuolar translocation sequence (PEXEL/VTS) that is characterized by the consensus sequence RX(L/I)X(D/E/Q). These signals are predicted to facilitate the transport of proteins into the PV (using their recessed, or N-terminal, signal sequences) and translocation across the PVM (using their PEXEL/VTS motifs) (5, 23, 29, 34). However, a subset of the exported proteins lack either one or both signal elements and may require novel targeting motifs for transport beyond the PPM (20, 43). A majority of the proteins enter the parasite secretory system via the endoplasmic reticulum (ER), where they are incorporated into ER-derived vesicles and then transported through the “unstacked” Golgi bodies to their final destinations (45, 48, 55, 56). Membrane-bound vesicular elements have been detected in the infected host cell cytosol, suggesting the existence of an extraparasitic vesicle-mediated transport process in malaria parasites (22, 47, 52). How vesicle targeting is achieved in P. falciparum parasites remains elusive.
Vesicle targeting and fusion in eukaryotic cells involves proteins of the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) family (25, 41, 42, 44). SNAREs are “tail-anchored” proteins that function by forming complexes that bridge vesicle and target membranes during fusion (6, 7, 24). Distinct sets of SNARE proteins localize to different intracellular transport pathways using processes that are not well understood. Increasing evidence suggests that the N-terminal regions of SNARE proteins contain signals required for their subcellular localization (4, 31, 53). These N-terminal regions include the three-helical Habc bundles of syntaxin SNAREs and the “profilin-like” folds of long VAMPs (vesicle-associated membrane proteins), also known as longin domains (7, 17, 33, 40, 46). The Sec22 gene products in mammals and yeast are longin domain-containing SNAREs that cycle between the ER and Golgi compartments (3, 19, 31, 32). We have identified a Sec22 ortholog in P. falciparum (PfSec22) that contains a PEXEL/VTS sequence insertion between the α2 and α3 segments of the longin domain preceded by a stretch of hydrophobic residues that spans a region between the β5 and α2 segments (2). In this study, we examined the distribution of PfSec22 in P. falciparum-infected erythrocytes and investigated the role of the atypical longin domain in its steady-state localization. Our data show that the P. falciparum ortholog of Sec22 partially associates with noncanonical destinations (tubovesicular network and intraerythrocytic vesicles) in the infected erythrocytes and that the N-terminal longin domain exhibits a dual function, mediating ER-to-Golgi apparatus trafficking, as well as retrieval from the Golgi apparatus.
MATERIALS AND METHODS
Plasmid constructs and transfection.
Transfection plasmids were designed to express wild-type or mutant green fluorescent protein (GFP)-tagged Sec22 proteins under the control of their upstream homologous promoter sequences. To achieve this, forward (5′-CCGGGGCCCTTTCCCTTCCCCGAAA-3′) and reverse (5′-CGGCCTAGGTGTTCTTTTTTTATTATTTTCTTCT-3′) primers were designed to amplify a 995-bp segment of the 1,781-bp intergenic region (5′ untranslated region) between PFC0890w and PFC0886w by PCR using P. falciparum 3D7 genomic DNA. The amplified sequence was ligated into the pGEMT-Easy vector (Promega) and subsequently subcloned into the PspoMI/AvrII sites of the previously generated construct pDC2-cam-GFP-PfSec22 (2, 30). This replaced the cam promoter with the 0.995-kb 5′-untranslated-region sequence of PfSec22 to obtain the construct pDC2-0.995. The resulting construct expressed full-length PfSec22 proteins with GFP fused to its N terminus (GFP-PfSec22). DNAs encoding PfSec22 mutant proteins were also generated by PCR and subcloned into the BglII and XhoI sites of the pDC-0.995vector. The primer sets, 5′-GGAAGATCTATGTGCGATGTAGTATTACTT-3′ and 5′-CCGCTCGAGTTATTTTAGATTTAATACCCTTGA-3′ or 5′-GGATCCAGCTTTTTTATTTTTAAACGAT-3′ and 5′-CCGCTCGAGTTAAAAATAATTTTTAAAAATTATAATT-3′ were used to amplify PfSec22 gene fragments to construct the GFP-PfSec22Δ198-221 and the GFP-PfSec22Δ1-78 fusion genes, respectively. DNA encoding amino acids 1 to 58 of the full-length Sec22 protein was also amplified using the primers 5′-GGAAGATCTATGTGCGATGTAGTATTACTT-3′ and 5′-GGATCCAAAATGGTAATTAAAATTGTTAG-3′ and subsequently ligated via the BamHI site to the C-terminal fragment of PfSec22 (GFP-PfSec22Δ1-78) cloned in a pGEMT-Easy vector. The ligated fragments were then subcloned into the pDC2-0.995 vector to obtain the GFP-PfSec22Δ58-78 mutant construct. To generate plasmids with GFP appended to the C terminus of PfSec22 (PfSec22-GFP), the full-length sequence was amplified using the primers 5′-CCTAGGATGTGCGATGTAGTATTACTT-3′ and 5′-AGATCTAAAATAATTTTTAAAAATTATAATTAAT-3′ and subsequently cloned into the AvrII/BglII sites of the pDC2-0.995 vector. The GFP-PfSec22Δ198-221.PEXEL(R>A) construct was generated from the GFP-PfSec22Δ198-221 mutant plasmid by site-directed mutagenesis of the PEXEL motif-specific arginine, replacing it with an alanine codon. All constructs were confirmed by sequencing prior to transfection of P. falciparum 3D7 parasites. Ring stage parasites were transfected by electroporation (Bio-Rad Gene Pulser II at 0.31 kV, 950 μF, and maximum capacitance) using 100 μg of purified plasmid (Qiagen Maxiprep kit). Positive selection for transfected parasites was achieved using 2.5 nM WR99210 as previously described (2, 16).
Anti-peptide antibodies and immunoblot analyses.
The decapeptide YKDPRSNIAI, corresponding to residues 131 to 140 of PfSec22, was synthesized (Genscript) and conjugated to keyhole limpet hemacyanin following the manufacturer's instructions (Pierce). Antisera were raised in rabbits against the conjugated peptide (Harlan Bioproducts for Science, Inc.) and affinity purified by column chromatography using peptide-conjugated agarose beads (Pierce). The reactivities of these antibodies against the endogenous and/or GFP-tagged PfSec22 proteins were analyzed by standard Western blotting techniques using a Supersignal West Femto detection kit (Pierce).
Immunofluorescence and confocal microscopy.
Both live and fixed cells were examined using a laser scanning confocal microscope (LSM 510; Carl Zeiss). The excitation/emission spectra settings were 488/505 nm for GFP, 543/555 for Alexa Fluor 555-conjugated antibodies, or 543/594 nm for Alexa Fluor 594 conjugates. To image live parasites, the cells were mounted under a coverslip in 50% glycerol and observed within 15 min of removal from cultures. Immunofluorescence assays were performed in suspension as previously described by Tonkin et al. (51). Rabbit anti-PfErd2 (MRA-1) and rat anti-PfBip (MRA-19) antibodies were obtained from the Malaria Research and Reference Resource Center and were each used at a dilution of 1 in 1,000. The purified rabbit anti-PfSec22 antibodies were also used at a 1 in 500 dilution. Secondary antibodies consisted of goat anti-rabbit Alexa Fluor 488, goat anti-rabbit Alexa Fluor 555, or goat anti-rat Alexa Fluor 594 (Molecular Probes), each used at a dilution of 1 in 1,000.
BFA treatment.
To investigate the effects of brefeldin A (BFA) on the steady-state location of PfSec22 and its mutant proteins, GFP-expressing cells were cultured in the presence of the agent at 5 μg/ml for 3 h (14) and then examined live as described above. Control experiments consisted of the same culture samples prepared in equivalent amounts (0.5% methanol in RPMI medium) of the drug solvent.
Subcellular fractionation analyses. (i) Freeze/thaw fractionation of soluble and membrane-associated proteins.
P. falciparum 3D7 parasites were purified by saponin treatment and resuspended in a Tris-buffered saline (TBS) buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl) containing a cocktail of protease inhibitors (Roche). The cells were subjected to five cycles of freezing and thawing in liquid nitrogen, followed by a brief sonication for 15 s to release both the cytosolic and the luminal proteins. The disrupted cells were clarified by centrifugation at 100,000 × g for 1 h at 4°C to separate the soluble proteins from the membrane-associated fractions. The pellet sample was then normalized with the TBS buffer to the volume of the supernatant, and equivalent volumes were analyzed by Western blotting.
(ii) Alkaline extraction of peripheral from integral membrane proteins.
Membrane pellets were prepared as described above and resuspended in 3 volumes of 0.1 M Na2CO3, pH 11. The suspension was mixed by rotation at 4°C for 30 min, followed by centrifugation as described above to separate the peripheral membrane proteins in the supernatant from the integral membrane protein in the pellet. The resulting pellet was normalized as described above, and equal volumes were analyzed by Western blotting.
(iii) Triton X-114 phase separation of hydrophobic from hydrophilic membrane proteins.
Integral membrane fractions were prepared by alkaline extraction as described above and solubilized using the Membrane Protein Extraction Reagent kit (Pierce). The solubilized fraction was clarified by centrifugation at 10,000 × g for 3 min at 4°C and then clouded in a 37°C water bath for 20 min. This was followed by centrifugation at 10,000 × g for 2 min at room temperature to separate the hydrophilic phase (top layer) from the hydrophobic protein phase (bottom layer). Equal volumes of the phase-separated samples were further diluted in Laemmli sample buffer and analyzed by Western blotting.
(iv) Sucrose density gradient fractionation of subcellular compartments.
Two hundred fifty to 300 mg of parasite pellets were resuspended in ice-cold TBS buffer containing protease inhibitor cocktail (Roche) in the presence of 2 mM EDTA or 5 mM MgCl2. The cells were disrupted by five freeze-thaw cycles and then cleared at 10,000 × g for 5 min at 4°C. The suspension was then adjusted to a 47% final sucrose concentration and deposited (in a final volume of 1.5 ml) at the bottom of a discontinuous sucrose gradient comprising 1 ml of 40% sucrose, 1 ml of 35% sucrose, 0.75 ml of 25% sucrose, and 0.75 ml of 20% sucrose. The gradient was centrifuged at 100,000 × g for 16 h at 4°C in a Beckman SW50.1 rotor and fractionated into 0.5-ml aliquots by piercing the bottom of the tube using a fraction recovery system (Beckman). Equal volumes of each fraction were analyzed by Western blotting using anti-PfSec22, anti-PfErd2, or anti-PfBip antibodies diluted 1 in 1,000.
Cryo-immunoelectron microscopy.
P. falciparum-infected erythrocytes were enriched by Percoll density centrifugation and fixed for 30 min at room temperature in 4% paraformaldehyde/0.1% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M phosphate buffer, pH 7.2 (Electron Microscopy Sciences). Infected red blood cells were washed three times with phosphate buffer and pelleted into 10% gelatin in phosphate buffer and infused with 2.3 M sucrose in phosphate buffer. The cryoprotected pellets were frozen in liquid nitrogen and cryotransferred to a Leica EMFCS chamber. Ultrathin cryosections were obtained at −120°C on a Leica Ultracut UCT using a Drukker ultramicrotome diamond knife. The resulting ribbons of frozen sections were collected onto carbon and Formvar substrates mounted on 300-mesh nickel grids. The grids were floated on drops of BD JL8 anti-GFP (diluted 1:100 in 0.05 M phosphate buffer containing 1% bovine serum albumin) at 5°C overnight and then incubated at room temperature for 1 h with a 1:50 dilution of 12-nm colloidal-gold-conjugated Affinipure goat anti-mouse immunoglobulin G/immunoglobulin M (Jackson ImmunoResearch Laboratories) in 0.05 M phosphate buffer containing 1% bovine serum albumin. The grids were fixed with 1% glutaraldehyde and then embedded in 1% methylcellulose and 2.5% uranyl acetate prior to examination in an FEI Technai 12 transmission electron microscope. The images were captured using an AMT XR111 digital camera.
RESULTS
Steady-state locations of endogenous PfSec22 protein.
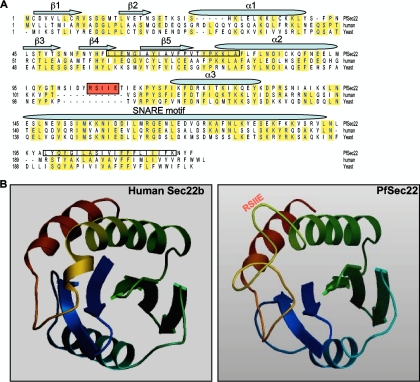
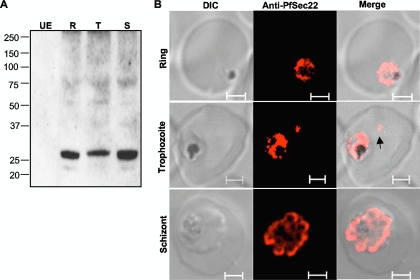
Sequence analysis of the Sec22 gene product in P. falciparum revealed a significant divergence from orthologs in yeast and mammals (overall identities of 26.2% and 30.8%, respectively) (1). The N-terminal longin domain of PfSec22 contains a bipartite signal sequence typical of proteins that are exported into the host cell compartment of P. falciparum-infected erythrocytes. These features include a PEXEL-like motif (105RSIIE109) located in an unusual loop-like region between the α2 and α3 segments of the longin domain and an N-terminal hydrophobic segment that is predicted (with a prediction probability ratio of 0.8192 [http://bioserve.latrobe.edu.au/cgi-bin/pfsigseq.py]) to function as a “recessed” signal sequence capable of targeting the protein for export beyond the PPM (Fig. 1A and B). We have previously shown, however, that expression of a GFP-tagged version of PfSec22 under the control of a heterologous cam promoter results in partial association of the protein with the parasite ER and Golgi apparatus, but not with the host cell compartment (2). To investigate whether overexpression of the protein in transgenic parasites had any influence on its steady-state dynamics, we generated anti-peptide antibodies that were used to detect the endogenous proteins by immunofluorescence microscopy and by Western blot analyses of cellular fractions. As shown in Fig. 2A, the affinity-purified antibodies specifically recognized a 26-kDa protein in cell extracts from ring, trophozoite, and schizont stage parasites, corresponding to the predicted molecular mass of the full-length PfSec22 protein. Expression of PfSec22 protein in all the asexual life cycle stages is consistent with the gene transcription profile (shown at http://www.PlasmoDB.org). By immunofluorescence analyses, we observed that the majority of the PfSec22 signal localized to membrane-like compartments inside the parasite cytoplasm. Consistently, albeit in a small proportion of cells (5.2% of 308 trophozoite-infected erythrocytes), we observed specific staining of isolated vesicular structures in the host cell cytoplasm using the anti-PfSec22 antibodies (Fig. 2B). Compared with the ER marker PfBip (P. falciparum binding protein), PfSec22 was observed to often be associated with the ER (Fig. 3A), consistent with a potential role in ER-to-Golgi apparatus transport in the malaria parasite. A similar coimmunofluorescence assay of the association of PfSec22 with the Golgi apparatus was precluded because the available anti-PfErd2 and anti-PfSec22 antibodies originated from the same animal species.
FIG. 1.
Sequence features of PfSec22. (A) Sequence alignment of PfSec22 with its homologues in humans and yeast. Identical amino acids are highlighted in yellow, while the PEXEL-like motif is shaded in red. The five beta strands (β1 to β5) and three alpha helix (α1 to α3) structures of the longin domain are indicated. The predicted hydrophobic segments (N-terminal and C-terminal hydrophobic segments) are boxed. PfSec22 and human Sec22b are 30.9% identical; PfSec22 and yeast Sec22p are 26.2% identical. In comparison, the human and yeast Sec22 polypeptides are 37.2% identical. (B) Ribbon structures of human Sec22b and PfSec22 longin domains showing the locations of the PEXEL-like motif (RSIIE). The PfSec22 structure was modeled with SWISS-MODEL 8.05 using the human Sec22b sequence as a template.
FIG. 2.
Expression analyses of PfSec22. Antibodies were generated against a peptide sequence in the nonconserved region of PfSec22 and affinity purified as described in Materials and Methods. (A) Immunoblot analysis of parasite extracts showing expression of endogenous PfSec22 in ring (R), trophozoite (T), and schizont (S) stage parasites. Absence of antibody reaction with the uninfected erythrocyte lysate (UE) indicates high specificity of the antibodies for the parasite protein. (B) Immunofluorescence microscopy of fixed cells using anti-PfSec22 antibodies and goat anti-rabbit Alexa Fluor 555 secondary antibodies. An intense ring of PfSec22 fluorescence is visible in ring- and trophozoite-infected cells. Isolated foci of PfSec22 fluorescence (arrow) are also detected in the host cell compartment in trophozoite-infected cells, suggesting export of PfSec22 into the erythrocyte cytosol. Scale bars, 2 μm.
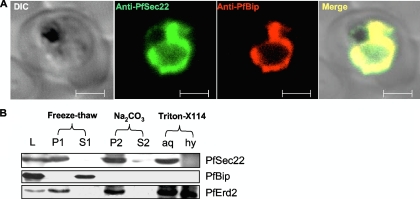
FIG. 3.
Coimmunolocalization and membrane association of endogenous PfSec22. (A) The association of PfSec22 with the parasite ER was investigated by coimmunofluorescence assays using rabbit anti-PfSec22 and rat anti-PfBip antibodies, as indicated. Both proteins colocalized significantly within the parasite. Scale bars, 2 μm. (B) Membrane association of PfSec22. Saponin-purified parasites were lysed by freeze-thaw/sonication and then centrifuged to separate the membrane-anchored (P1) from the soluble cytosolic and luminal (S1) proteins. Integral membrane proteins (P2) were separated from the peripheral membrane proteins (S2) by alkaline extraction with 0.1 M Na2CO3 solution at pH 11. The solubility profiles of the integral membrane proteins were further analyzed by Triton X-114 extraction to separate the hydrophilic proteins in the aqueous (aq) phase from the hydrophobic proteins (hy) in the detergent phase. Normalized volumes of the sample pairs were loaded into each well and immunoblotted with antibodies against the PfSec22 protein, the soluble protein PfBip, or the integral protein marker PfErd2. Thirty-five micrograms of the freeze-thaw lysate was loaded in lane L.
To establish that PfSec22 is membrane anchored in the parasite, we investigated its membrane properties by (i) freeze-thaw fractionation of membrane-associated proteins from soluble fractions, (ii) alkaline extraction of peripheral membrane proteins from integral transmembrane proteins, and (iii) Triton X-114 solubilization of integral membrane proteins, followed by immunodetection. The ER-restricted luminal chaperone PfBip was used as a marker for the soluble non-membrane-associated proteins, whereas the presence of the KDEL receptor PfErd2 (the P. falciparum homologue of ER retention-deficient mutant 2) in the fractions marked the membrane-associated proteins. As shown in Fig. 3B, the endogenous PfSec22 protein partitioned alongside PfErd2 in the membrane fractions and was efficiently solubilized into the hydrophilic phase after detergent extraction using Triton X-114. PfErd2 was only partly solubilized by the detergent, consistent with its topological function as a multipass membrane protein containing up to seven transmembrane domains (14). These results suggest that PfSec22 is an integral membrane protein, consistent with a requirement in vesicle trafficking.
Localization of PfSec22 in transgenic parasites.
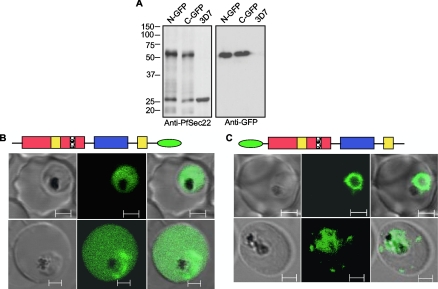
To better understand the intracellular dynamics of this unusual Sec22 gene product, we generated a transgenic cell line expressing the GFP-tagged proteins under the control of the endogenous PfSec22 promoter sequence. Additionally, to ensure that a suitable tagging approach was being employed, we first investigated the effect of the GFP tag. Transgenic parasites were developed that expressed the PfSec22 protein with GFP appended either to its C terminus (PfSec22-GFP) or to its N terminus (GFP-PfSec22). We confirmed the expression of each fusion protein by immunoblot analyses using either anti-PfSec22 or anti-GFP antibodies (Fig. 4A). As indicated in Fig. 4A, two protein bands corresponding to the expected masses of the GFP-tagged proteins (∼54 kDa) and the endogenous protein (26 kDa) were detected in each transgenic cell lysate using the anti-PfSec22 antibodies. In contrast, only the 54-kDa bands were detected in these extracts using the anti-GFP antibodies. These data established that full-length proteins were expressed in the respective cell lines and that PfSec22 is not processed at the N terminus, as has been reported for most PEXEL-containing proteins (8).
FIG. 4.
Live-cell imaging of GFP-tagged PfSec22 chimeras. (A) Immunoblot analyses using anti-PfSec22 antibodies (left blot) or monoclonal anti-GFP antibodies (right blot) confirmed expression of the N-terminal GFP-tagged PfSec22 (N-GFP) and the C-terminal GFP-tagged PfSec22 (C-GFP) proteins (∼54 kDa) in the respective transgenic cell lines, but not in untransfected parasites (3D7). The anti-PfSec22 antibodies also detected an ∼26-kDa protein that corresponded to the untagged PfSec22 protein in whole-cell extracts from all three cell lines. (B) Live-cell imaging of parasites expressing the C-terminal GFP-tagged PfSec22 (PfSec22-GFP) showing diffuse localization of the protein throughout the parasite cytoplasm and occasionally in the host cell compartment. (C) Confocal micrographs showing localization of the N-terminal GFP-tagged protein (GFP-PfSec22) in early- trophozoite (top row) and mid-trophozoite (bottom row) stage parasites. In addition to the ER-like profiles, GFP-PfSec22 associates with tubovesicular elements in the infected host cell. The micrographs (left to right) represent differential interference contrast, GFP fluorescence, and a merge of the two. Scale bars, 2 μm.
Live-cell imaging of GFP fluorescence revealed significant differences in the distribution of GFP-PfSec22 and PfSec22-GFP (Fig. 4B and C, respectively). In transgenic parasites expressing the C-terminally tagged protein, the GFP fluorescence was observed predominantly in the parasite cytosol (Fig. 4B). In some trophozoite-infected cells, representing 9.3% of 302 trophozoite-infected erythrocytes, the GFP fluorescence was also observed inside the erythrocyte cytosol (Fig. 4B, bottom row). In comparison, the N-terminally tagged protein (GFP-PfSec22) localized predominantly to membranous structures located inside the parasite cytoplasm (Fig. 4C). Similar to the untagged PfSec22 protein, the GFP-PfSec22 protein was also detected inside the host cell compartment in a small proportion (approximately 5%) of trophozoite-infected cells. There, it associated with various tubovesicular extensions (Fig. 4C, bottom row) and also with mobile vesicular structures (data not shown). The cellular locations of GFP-PfSec22 were further investigated at the ultrastructural level using monoclonal anti-GFP antibodies and by immunofluorescence analyses (Fig. 5 and 6, respectively). Immunogold labeling of ultrathin cryosections confirmed that GFP-PfSec22 localized predominantly to the parasite ER (Fig. 5A). Membrane vesicles were also labeled in the parasite cytoplasm (Fig. 5A) and in the erythrocyte cytosol of some trophozoite-infected cells (Fig. 5B and C). Together, these findings suggest that PfSec22 might function as a vesicle-associated SNARE (v-SNARE) in the malaria parasite. Further attempts to fractionate the exported proteins by limited permeabilization of the host cell membrane were unsuccessful, presumably due to its membrane localization and/or low levels in the erythrocyte cytosol.
FIG. 5.
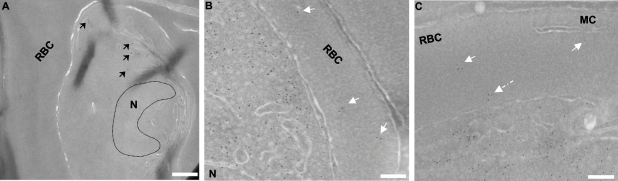
Immunoelectron micrographs depicting the association of GFP-PfSec22 with transition vesicles and tubovesicular elements. Ultrathin cryosections of the GFP-PfSec22-expressing cells were probed with monoclonal anti-GFP antibodies, followed by immunogold detection using gold (12 nm)-labeled anti-mouse secondary antibodies. (A) Association of GFP-PfSec22 with ER-derived transition vesicles (arrows). (B) Association of GFP-PfSec22 with membrane-limited vesicles (arrows) in the host cell compartment. (C) Association of GFP-PfSec22 with the TVN-like extension (dashed arrow) and membrane-bound vesicles (solid arrows) in the infected erythrocyte cytosol. N, nucleus; MC, Maurer's cleft; RBC, red blood cell cytosol. Scale bars, 500 nm (A) and 100 nm (B and C).
FIG. 6.
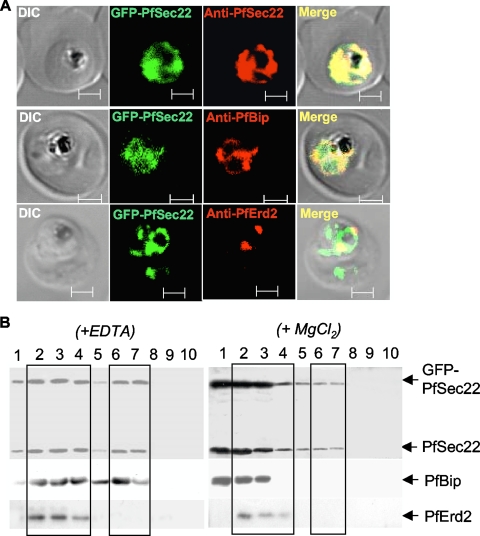
Colocalization of GFP-PfSec22 with ER and cis-Golgi markers. (A) Immunoflorescence micrographs indicating colocalization of the GFP-PfSec22 and anti-PfSec22 signals in transgenic parasites (top row) and colocalization of GFP-PfSec22 with the ER marker PfBip (middle row) and the Golgi apparatus marker PfErd2 (bottom row). Scale bars, 2 μm. (B) Sucrose density gradient fractionation of GFP-PfSec22 and untagged PfSec22 in the presence of 2 mM EDTA (left) or in the presence of 5 mM MgCl2 (right). The numbers represent the fraction numbers, collected from bottom (fraction 1) to top (fraction 10), of a discontinuous sucrose density gradient consisting of 1.5 ml of 47%, 1 ml of 40%, 1 ml of 35%, 0.75 ml of 25%, and 0.75 ml of 20% sucrose. GFP-PfSec22 and the untagged protein both partially redistributed to the high-density fractions (fractions 1 to 4) alongside PfBip in the presence of MgCl2, suggestive of PfSec22 localization to the ER. DIC, differential interference contrast.
To validate the use of GFP-tagged proteins in our study, we compared the GFP and PfSec22 signals by immunofluorescence analysis using the anti-PfSec22 antibodies. As shown in Fig. 6A, top row, the anti-PfSec22 signal in the GFP-PfSec22-expressing parasites was limited to the GFP fluorescence structures, suggesting that the untagged wild-type protein also localized to the same cytoplasmic compartments as the GFP-tagged chimera. GFP-PfSec22 also colocalized significantly with PfBip (Fig. 6A, middle row) and PfErd2 (Fig. 6A, bottom row), suggesting that PfSec22 is predominantly a v-SNARE of the parasite ER/Golgi apparatus pathway. As an alternative approach to confirm the association of GFP-PfSec22 with the ER and Golgi apparatus, we investigated its cofractionation with (i) the untagged PfSec22 protein, (ii) the ER marker PfBip, and (iii) the Golgi apparatus marker PfErd2 by sucrose density gradient ultracentrifugation and immunoblot analyses (Fig. 6B). To overcome common difficulties in resolving the parasite ER from other membrane-bound compartments, the cofractionation experiments were done in the presence of the chelating agent EDTA or in the presence of the rough-ER-stabilizing agent MgCl2 (11, 28). By this approach, a redistribution of the PfSec22 proteins alongside the ER marker PfBip from a low-density fraction (in the presence of EDTA) to a high-density fraction (in the presence of MgCl2) suggested its association with the rough ER. As shown in Fig. 6B, the untagged (PfSec22) and the tagged (GFP-PfSec22) proteins similarly redistributed alongside PfBip to the high-sucrose-density fractions following treatment with MgCl2. However, only a limited amount of these PfSec22 proteins shifted to the high-density fractions compared with PfBip, which was completely redistributed to the high-density fractions in the presence of the Mg2+ cations. These results also suggest that PfSec22 associates with non-ER structures, presumably including transport vesicles, as revealed by our electron microscopy studies (Fig. 5). Neither EDTA nor MgCl2 affected the buoyant density of the Golgi apparatus in our gradients, as was evident from the distribution of PfErd2. Taken together, our data indicate that PfSec22 is predominantly a v-SNARE of the parasite ER/Golgi apparatus interface and that this SNARE protein might also play a limited role in the host cell remodeling process, specifically in late-trophozoite-infected cells.
Trafficking determinants for PfSec22.
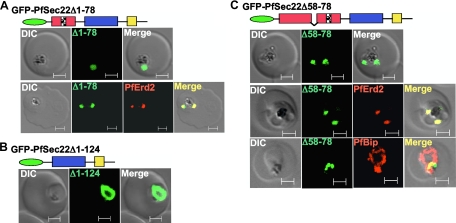
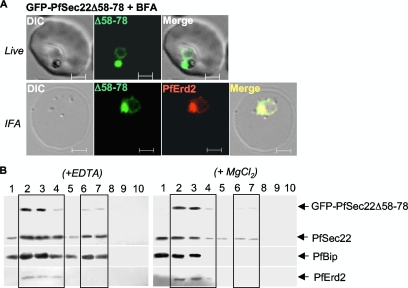
In an attempt to decipher the signals that determine the steady-state localization of PfSec22 in the parasite, we investigated the roles of the atypical longin domain, the two hydrophobic segments, and the PEXEL/VTS-like sequence. Transgenic parasites that expressed various N-terminally tagged PfSec22 mutants were generated. As shown in Fig. 7A, truncation of the longin domain at position 78 (the GFP-PfSec22Δ1-78 mutant) dramatically shifted its steady-state accumulation from the ER to the Golgi apparatus, based on its colocalization with PfErd2 (Fig. 7A, bottom row). These findings suggest that GFP-PfSec22Δ1-78 presumably has a preference for the parasite Golgi apparatus and that residues 1 to 78 might encode signals required for ER retrieval of PfSec22. Interestingly, deletion of the entire longin domain at position 124 (GFP-PfSec22Δ1-124) resulted in an ER localization of the protein (Fig. 7B and colocalization data not shown), suggesting that this mutant was not exported beyond the ER compartment. In contrast, a minimized deletion of the N-terminal hydrophobic segment ( the GFP-PfSec22Δ58-78 mutant) resulted in Golgi apparatus localization (Fig. 7C). Similar to the GFP-PfSec22Δ1-78 mutant, this GFP-PfSec22Δ58-78 mutant colocalized significantly with the Golgi apparatus marker PfErd2, but not with the ER marker PfBip (Fig. 7C, middle and bottom rows, respectively). The Golgi apparatus targeting of this N-terminal hydrophobic domain mutant was sensitive to BFA treatment (Fig. 8A), suggesting its association with ER/Golgi apparatus transport vesicles. As shown in Fig. 8A, bottom row, BFA induced a partial redistribution of both the GFP-PfSec22Δ58-78 mutant and PfErd2 to the ER, a phenomenon typical of most Golgi apparatus-localized proteins (14, 37). We confirmed the differential localization of GFP-PfSec22Δ58-78 to the Golgi apparatus by discontinuous sucrose density ultracentrifugation in the presence of EDTA or MgCl2 (Fig. 8B). Compared to the endogenous wild-type protein, which redistributed alongside PfBip to the high-density fractions in the presence of MgCl2, the buoyant density of the GFP-PfSec22Δ58-78-associated compartments did not change in the presence of EDTA or MgCl2 (Fig. 8B). A small proportion of the mutant protein remained in the low-density zone, consistent with an association with transport vesicles and/or other lipid-rich compartments. Together, our data suggest that the ER exit of PfSec22 is independent of residues 1 to 78 of the longin domain and that the N-terminal hydrophobic region (amino acids 58 to 78) is required for the retrograde transport of PfSec22 back to the ER.
FIG. 7.
Role of the atypical longin domain in PfSec22 trafficking. (A and B) Transgenic parasites were generated expressing PfSec22 proteins that lacked amino acids 1 to 78 (GFP-PfSec22Δ1-78) (A) or amino acids 1 to 124 (GFP-PfSec22Δ1-124), corresponding to the entire longin domain (B), or that lacked residues 58 to 78 (GFP-PfSec22Δ58-78), corresponding to the 21-amino-acid N-terminal hydrophobic segment (C). Both the GFP-PfSec22Δ1-78 and the GFP-PfSec22Δ58-78 mutants colocalized with PfErd2, but not with the ER marker PfBip, suggesting Golgi apparatus retention of the N-terminal hydrophobic domain mutants. In contrast, the GFP-PfSec22Δ1-124 mutant was retained in the ER (B). Scale bars, 2 μm. DIC, differential interference contrast.
FIG. 8.
Effect of BFA and sucrose density gradient fractionation of GFP-PfSec22Δ58-78. (A) Treatment of the GFP-PfSec22Δ58-78-expressing parasites with BFA resulted in partial redistribution of the mutant protein to the ER (top row), similarly to the Golgi apparatus-localized PfErd2 protein (bottom row). Treatment of the parasites with the drug solvent (0.5% methanol in RPMI medium) did not inhibit GFP-PfSec22Δ58-78 transport to the Golgi apparatus (data not shown). Scale bars, 2 μm. IFA, immunofluorescence assay. (B) Sucrose density gradient fractionation and coimmunodetection of GFP-PfSec22Δ58-78 with wild-type PfSec22 protein, PfBip, and PfErd2 antigens. Compared with the untagged wild-type protein (PfSec22), which redistributed alongside PfBip in the presence of MgCl2,the GFP-PfSec22Δ58-78 mutant was consistently detected in the high-density fractions in the presence of EDTA (left) or MgCl2 (right). The numbers represent fraction numbers collected from the bottom (fraction 1) to the top (fraction 10) of a discontinuous sucrose density gradient consisting of 1.5 ml of 47%, 1 ml of 40%, 1 ml of 35%, 0.75 ml of 25%, and 0.75 ml of 20% sucrose. A small amount of GFP-PfSec22Δ58-78 was also detected in the low-density fractions, suggesting an association with lipid-rich structures that might represent transport vesicles. DIC, differential interference contrast.
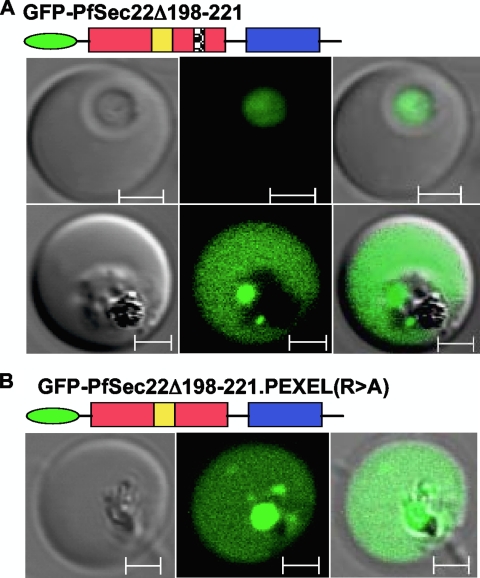
To establish the role of the C-terminal hydrophobic segment, we generated a transgenic cell line expressing the deletion mutant GFP-PfSec22Δ198-221. Deletion of this hydrophobic segment (amino acids 198 to 221) resulted in a cytosolic protein that occasionally was also released into the erythrocyte cytosol in a pattern similar to that of the C-terminally tagged full-length protein (compare Fig. 9A and Fig. 4B). These results support a role of the C-terminal hydrophobic segment in membrane anchorage and suggest that the occasional export of PfSec22 into the infected erythrocyte may involve processes independent of its membrane insertion. Such processes, however, are not likely to involve a canonical PEXEL/VTS signal, given that PfSec22 does not exhibit N-terminal cleavage at this motif. Furthermore, mutation of the critical arginine residue using our C-terminal hydrophobic domain mutant failed to ablate the protein export (Fig. 9B).
FIG. 9.
Roles of the C-terminal hydrophobic domain and PEXEL motif. (A) Deletion of the C-terminal hydrophobic domain (GFP-PfSec22Δ198-221) resulted in diffuse localization within the parasite cytoplasm and in the infected host cell in a pattern similar to that of the C-terminally tagged full-length protein (Fig. 4B). (B) Replacement of the PEXEL arginine with alanine (i.e., RSIIE to ASIIE) did not inhibit export of the soluble GFP-PfSec22Δ198-221 protein to the erythrocyte cytoplasm. These results suggest that the C-terminal hydrophobic domain plays an essential role in membrane anchorage of PfSec22 and that the export process of PfSec22 is independent of its membrane integration and the PEXEL-like motif. Scale bars, 2 μm.
DISCUSSION
The N-terminal longin domains of SNARE proteins have been proposed to mediate the differential localization of these proteins to intracellular compartments, and in some cases, as for ykt6 gene products, to regulate the SNARE activity (18, 21, 32, 35, 39, 53). The longin domain of PfSec22 exhibits significant differences from yeast and human orthologs, which might influence its cellular locations and trafficking mechanisms. These features include a decapeptide sequence insertion within an unusual loop-like region of the longin domain (Fig. 1A) and an N-terminal hydrophobic segment predicted to function as a recessed signal sequence. We examined the subcellular distribution of PfSec22 and investigated the roles of the atypical longin domain and the C-terminal hydrophobic segment in its steady-state localization. Consistent with the subcellular locations of Sec22 proteins in model organisms (3, 10, 26, 38, 54), PfSec22 localized predominantly to the ER-Golgi apparatus interface in P. falciparum. However, we consistently detected the GFP-tagged and the untagged proteins in tubovesicular structures, and in single vesicular compartments, inside the infected host cell (Fig. 2B, 4B, and 6A). These findings are consistent with the findings of Trelka et al. (52), in which single vesicles, or vesicular aggregates of various sizes, were observed in trophozoite-infected erythrocytes. Until now, the significance of these less frequently occurring vesicular structures in the host cell compartment has not been determined. In the previous report, treatment of the parasites with aluminum tetrafluoride, an activator of GTP-binding proteins, resulted in the accumulation of electron-dense vesicles inside the parasitized erythrocyte cytosol. A similar aluminum fluoride treatment of the GFP-PfSec22-expressing parasites in our study, however, did not accumulate the GFP fluorescence in the host cell compartment (data not shown), suggesting that the PfSec22-associated structures might represent a distinct subpopulation of the extraparasitic vesicles. It was not clear in our studies whether export of the Sec22 gene product in the malaria parasite was a direct consequence of the atypical longin domain or due to the unusual organization of the parasite secretory system.
To gain insights into the PfSec22 targeting signals, as a model for SNARE protein targeting in P. falciparum, we investigated the role of the atypical longin domain. Our data suggest that this domain regulates the steady-state localization of PfSec22 to the parasite ER/Golgi apparatus interface and its putative transport to the erythrocyte cytoplasm. We found that ER exit of PfSec22 requires residues within the α3 helix, but with no absolute requirement for the entire longin domain. These findings are in contrast with studies of yeast, wherein truncation of the Sec22p longin domain resulted in ER retention (31), thus suggesting a mechanistic difference in the PfSec22 export process. It is well established in yeast and mammals that the packaging of Sec22 into COPII vesicles involves interaction between the α3 segment of the longin domain and a preformed Sec23/Sec24 complex (31, 32). Two conserved residues of the α3 segment, I113 and D116 in Sec22b (corresponding to I118 and D121 in PfSec22), are required for this interaction and therefore for Sec22 exit from the ER (32). Exposure of the α3 residues for binding to the Sec23/Sec24 complex is thought to be induced by a conformational epitope that is formed by interaction of an “NIE” motif of the SNARE domain and the α1 and β3 segments (32). Presumably, truncation of the PfSec22 longin domain at position 78 was sufficient to trigger exit from the ER. Alternatively, ER export of the Golgi apparatus-localized mutant proteins (GFP-PfSec22Δ1-78 and GFP-PfSec22Δ58-78) might have involved interactions with other endogenous proteins, including the untagged PfSec22 protein. We interpreted Golgi apparatus retention of the N-terminal hydrophobic domain mutants as a defect in the recycling process of these proteins. In yeast, recycling of Sec22p requires interactions with the COPI budding complex and with the SNARE proteins Ufe1 and Sec20 (3). Expression of yeast Sec22p in sec20-1 or ufe1-1 mutant cells or in the COPI mutants sec21-1 and sec27-1 results in accumulation of the protein in the Golgi apparatus and a lack of ER staining by anti-Sec22 antibodies (3). In support of a Golgi apparatus retention of the PfSec22 mutant protein (GFP-PfSec22Δ58-78), treatment of parasites with BFA resulted in partial redistribution of the protein to the ER (Fig. 8A), a phenomenon typical of most Golgi apparatus-localized proteins, including the cis-Golgi marker PfErd2 (14). BFA is a GTPase inhibitor that disrupts vesicle trafficking in both the retrograde and the anterograde pathways, resulting in retention of Golgi apparatus-resident proteins in the ER (13, 27, 36, 37).
Taken together, our data indicate that the longin domain of PfSec22 exhibits both structural and functional differences from yeast and mammalian Sec22 proteins. The potential for these deviations is supported by both the lack of significant sequence identity between the longin domains and the unusual organization of the parasite ER and Golgi compartments. Our data suggest that the N-terminal hydrophobic segment (amino acids 58 to 78) plays an essential role in retrograde transport of this v-SNARE. To our knowledge, this study provides the first experimental evidence for the role of the Sec22 longin domain in transporter recycling, a process that is likely to involve interactions with other SNARE proteins and the COPI machinery. We have also shown in this study that the C-terminal hydrophobic domain of PfSec22 is important for the membrane association of the protein and that the occasional export of PfSec22 into the host cell was independent of the PEXEL-like motif. In support of our views, PfSec22 is not processed at this motif, as has otherwise been reported for canonical PEXEL motif-containing proteins (8). Additionally, and in contrast with point mutation studies using fusion proteins that are efficiently exported to the erythrocyte cytosol in a PEXEL motif-dependent process (5), mutation of the PEXEL-like arginine of PfSec22 did not inhibit export of the C-terminal hydrophobic mutant (Fig. 9B). In light of the above-mentioned results and difficulties with the occasional export of PfSec22, it will now be important to identify all PfSec22-interacting partners and to characterize their subcellular locations.
Acknowledgments
We thank Marcus Lee of Columbia University for helpful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 2.Ayong, L., G. Pagnotti, A. B. Tobon, and D. Chakrabarti. 2007. Identification of Plasmodium falciparum family of SNAREs. Mol. Biochem. Parasitol. 152:113-122. [DOI] [PubMed] [Google Scholar]
- 3.Ballensiefen, W., D. Ossipov, and H. D. Schmitt. 1998. Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J. Cell Sci. 111:1507-1520. [DOI] [PubMed] [Google Scholar]
- 4.Banfield, D. K., M. J. Lewis, C. Rabouille, G. Warren, and H. R. Pelham. 1994. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J. Cell Biol. 127:357-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddey, J. A., R. L. Moritz, R. J. Simpson, and A. F. Cowman. 2009. Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10:285-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgese, N., S. Brambillasca, P. Soffientini, M. Yabal, and M. Makarow. 2003. Biogenesis of tail-anchored proteins. Biochem. Soc. Trans. 31:1238-1242. [DOI] [PubMed] [Google Scholar]
- 7.Brunger, A. T. 2005. Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys. 38:1-47. [DOI] [PubMed] [Google Scholar]
- 8.Chang, H. H., A. M. Falick, P. M. Carlton, J. W. Sedat, J. L. DeRisi, and M. A. Marletta. 2008. N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charpian, S., and J. M. Przyborski. 2008. Protein transport across the parasitophorous vacuole of Plasmodium falciparum: into the great wide open. Traffic 9:157-165. [DOI] [PubMed] [Google Scholar]
- 10.Chatre, L., F. Brandizzi, A. Hocquellet, C. Hawes, and P. Moreau. 2005. Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 139:1244-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chehab, E. W., O. R. Patharkar, and J. C. Cushman. 2007. Isolation and characterization of a novel v-SNARE family protein that interacts with a calcium-dependent protein kinase from the common ice plant, Mesembryanthemum crystallinum. Planta 225:783-799. [DOI] [PubMed] [Google Scholar]
- 12.Cooke, B. M., K. Lingelbach, L. H. Bannister, and L. Tilley. 2004. Protein trafficking in Plasmodium falciparum-infected red blood cells. Trends Parasitol. 20:581-589. [DOI] [PubMed] [Google Scholar]
- 13.Donaldson, J. G., D. Finazzi, and R. D. Klausner. 1992. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360:350-352. [DOI] [PubMed] [Google Scholar]
- 14.Elmendorf, H. G., and K. Haldar. 1993. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. EMBO J. 12:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epp, C., and K. Deitsch. 2006. Deciphering the export pathway of malaria surface proteins. Trends Parasitol. 22:401-404. [DOI] [PubMed] [Google Scholar]
- 16.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naude, K. W. Deitsch, X. Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippini, F., V. Rossi, T. Galli, A. Budillon, M. D'Urso, and M. D'Esposito. 2001. Longins: a new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem. Sci. 26:407-409. [DOI] [PubMed] [Google Scholar]
- 18.Fukasawa, M., O. Varlamov, W. S. Eng, T. H. Sollner, and J. E. Rothman. 2004. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc. Natl. Acad. Sci. USA 101:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, L. C., Jr., W. I. Weis, and R. H. Scheller. 2001. A novel snare N-terminal domain revealed by the crystal structure of Sec22b. J. Biol. Chem. 276:24203-24211. [DOI] [PubMed] [Google Scholar]
- 20.Haldar, K., B. U. Samuel, N. Mohandas, T. Harrison, and N. L. Hiller. 2001. Transport mechanisms in Plasmodium-infected erythrocytes: lipid rafts and a tubovesicular network. Int. J. Parasitol. 31:1393-1401. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa, H., S. Zinsser, Y. Rhee, E. O. Vik-Mo, S. Davanger, and J. C. Hay. 2003. Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell 14:698-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibbs, A. R., and A. J. Saul. 1994. Plasmodium falciparum: highly mobile small vesicles in the malaria-infected red blood cell cytoplasm. Exp. Parasitol. 79:260-269. [DOI] [PubMed] [Google Scholar]
- 23.Hiller, N. L., S. Bhattacharjee, C. van Ooij, K. Liolios, T. Harrison, C. Lopez-Estrano, and K. Haldar. 2004. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306:1934-1937. [DOI] [PubMed] [Google Scholar]
- 24.Hong, W. 2005. SNAREs and traffic. Biochim. Biophys. Acta 1744:120-144. [DOI] [PubMed] [Google Scholar]
- 25.Jahn, R., and R. H. Scheller. 2006. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631-643. [DOI] [PubMed] [Google Scholar]
- 26.Kissmehl, R., C. Schilde, T. Wassmer, C. Danzer, K. Nuehse, K. Lutter, and H. Plattner. 2007. Molecular identification of 26 syntaxin genes and their assignment to the different trafficking pathways in Paramecium. Traffic 8:523-542. [DOI] [PubMed] [Google Scholar]
- 27.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, D. J., P. B. Moore, and T. A. Steitz. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10:1366-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuepfer, E., M. Rug, and A. F. Cowman. 2005. Function of the plasmodium export element can be blocked by green fluorescent protein. Mol. Biochem. Parasitol. 142:258-262. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. C., P. A. Moura, E. A. Miller, and D. A. Fidock. 2008. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol. Microbiol. 68:1535-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Y., J. J. Flanagan, and C. Barlowe. 2004. Sec22p export from the endoplasmic reticulum is independent of SNARE pairing. J. Biol. Chem. 279:27225-27232. [DOI] [PubMed] [Google Scholar]
- 32.Mancias, J. D., and J. Goldberg. 2007. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell 26:403-414. [DOI] [PubMed] [Google Scholar]
- 33.Margittai, M., D. Fasshauer, R. Jahn, and R. Langen. 2003. The Habc domain and the SNARE core complex are connected by a highly flexible linker. Biochemistry 42:4009-4014. [DOI] [PubMed] [Google Scholar]
- 34.Marti, M., R. T. Good, M. Rug, E. Knuepfer, and A. F. Cowman. 2004. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306:1930-1933. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Arca, S., R. Rudge, M. Vacca, G. Raposo, J. Camonis, V. Proux-Gillardeaux, L. Daviet, E. Formstecher, A. Hamburger, F. Filippini, M. D'Esposito, and T. Galli. 2003. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA 100:9011-9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orci, L., A. Perrelet, M. Ravazzola, F. T. Wieland, R. Schekman, and J. E. Rothman. 1993. “BFA bodies”: a subcompartment of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 90:11089-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orci, L., M. Tagaya, M. Amherdt, A. Perrelet, J. G. Donaldson, J. Lippincott-Schwartz, R. D. Klausner, and J. E. Rothman. 1991. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell 64:1183-1195. [DOI] [PubMed] [Google Scholar]
- 38.Paek, I., L. Orci, M. Ravazzola, H. Erdjument-Bromage, M. Amherdt, P. Tempst, T. H. Sollner, and J. E. Rothman. 1997. ERS-24, a mammalian v-SNARE implicated in vesicle traffic between the ER and the Golgi. J. Cell Biol. 137:1017-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossi, V., D. K. Banfield, M. Vacca, L. E. Dietrich, C. Ungermann, M. D'Esposito, T. Galli, and F. Filippini. 2004. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 29:682-688. [DOI] [PubMed] [Google Scholar]
- 40.Rossi, V., R. Picco, M. Vacca, M. D'Esposito, M. D'Urso, T. Galli, and F. Filippini. 2004. VAMP subfamilies identified by specific R-SNARE motifs. Biol. Cell 96:251-256. [DOI] [PubMed] [Google Scholar]
- 41.Rothman, J. E. 1996. The protein machinery of vesicle budding and fusion. Protein Sci. 5:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothman, J. E. 2002. Lasker Basic Medical Research Award. The machinery and principles of vesicle transport in the cell. Nat. Med. 8:1059-1062. [DOI] [PubMed] [Google Scholar]
- 43.Saridaki, T., K. S. Frohlich, C. B. Breton, and M. Lanzer. 2008. Export of PfSBP1 to the Plasmodium falciparum Maurer's clefts. Traffic 10:137-152. [DOI] [PubMed] [Google Scholar]
- 44.Sollner, T. H., and J. E. Rothman. 1996. Molecular machinery mediating vesicle budding, docking and fusion. Experientia 52:1021-1025. [DOI] [PubMed] [Google Scholar]
- 45.Struck, N. S., S. Herrmann, I. Schmuck-Barkmann, S. de Souza Dias, S. Haase, A. L. Cabrera, M. Treeck, C. Bruns, C. Langer, A. F. Cowman, M. Marti, T. Spielmann, and T. W. Gilberger. 2008. Spatial dissection of the cis- and trans-Golgi compartments in the malaria parasite Plasmodium falciparum. Mol. Microbiol. 67:1320-1330. [DOI] [PubMed] [Google Scholar]
- 46.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395:347-353. [DOI] [PubMed] [Google Scholar]
- 47.Taraschi, T. F., M. O'Donnell, S. Martinez, T. Schneider, D. Trelka, V. M. Fowler, L. Tilley, and Y. Moriyama. 2003. Generation of an erythrocyte vesicle transport system by Plasmodium falciparum malaria parasites. Blood 102:3420-3426. [DOI] [PubMed] [Google Scholar]
- 48.Taraschi, T. F., D. Trelka, T. Schneider, and I. Matthews. 1998. Plasmodium falciparum: characterization of organelle migration during merozoite morphogenesis in asexual malaria infections. Exp. Parasitol. 88:184-193. [DOI] [PubMed] [Google Scholar]
- 49.Templeton, T. J., and K. W. Deitsch. 2005. Targeting malaria parasite proteins to the erythrocyte. Trends Parasitol. 21:399-402. [DOI] [PubMed] [Google Scholar]
- 50.Tonkin, C. J., J. A. Pearce, G. I. McFadden, and A. F. Cowman. 2006. Protein targeting to destinations of the secretory pathway in the malaria parasite Plasmodium falciparum. Curr. Opin. Microbiol. 9:381-387. [DOI] [PubMed] [Google Scholar]
- 51.Tonkin, C. J., G. G. van Dooren, T. P. Spurck, N. S. Struck, R. T. Good, E. Handman, A. F. Cowman, and G. I. McFadden. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137:13-21. [DOI] [PubMed] [Google Scholar]
- 52.Trelka, D. P., T. G. Schneider, J. C. Reeder, and T. F. Taraschi. 2000. Evidence for vesicle-mediated trafficking of parasite proteins to the host cell cytosol and erythrocyte surface membrane in Plasmodium falciparum infected erythrocytes. Mol. Biochem. Parasitol. 106:131-145. [DOI] [PubMed] [Google Scholar]
- 53.Uemura, T., M. H. Sato, and K. Takeyasu. 2005. The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett. 579:2842-2846. [DOI] [PubMed] [Google Scholar]
- 54.Uemura, T., T. Ueda, R. L. Ohniwa, A. Nakano, K. Takeyasu, and M. H. Sato. 2004. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29:49-65. [DOI] [PubMed] [Google Scholar]
- 55.van Dooren, G. G., M. Marti, C. J. Tonkin, L. M. Stimmler, A. F. Cowman, and G. I. McFadden. 2005. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol. Microbiol. 57:405-419. [DOI] [PubMed] [Google Scholar]
- 56.van Dooren, G. G., R. F. Waller, K. A. Joiner, D. S. Roos, and G. I. McFadden. 2000. Traffic jams: protein transport in Plasmodium falciparum. Parasitol. Today 16:421-427. [DOI] [PubMed] [Google Scholar]
- 57.Wickert, H., F. Wissing, K. T. Andrews, A. Stich, G. Krohne, and M. Lanzer. 2003. Evidence for trafficking of PfEMP1 to the surface of P. falciparum-infected erythrocytes via a complex membrane network. Eur. J. Cell Biol. 82:271-284. [DOI] [PubMed] [Google Scholar]