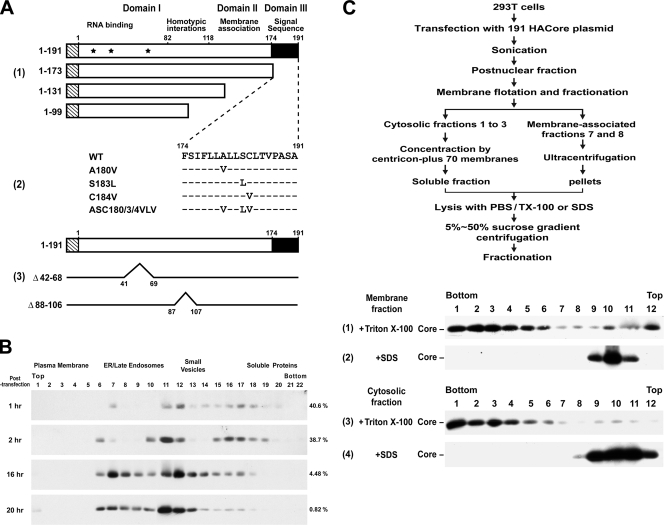
FIG. 1.
Construction of core plasmids and analyses of membrane-bound and -free core complexes. (A) pCAGGS-based plasmids encoding residues 1 to 191 of the genotype 1a H77 strain core protein and C-terminal-truncated mutants tagged with the influenza virus HA epitope at the N terminus, as shown by the striped rectangles, were constructed as described in Materials and Methods. Domain I, which contains three nuclear localization sequences (located at residues 6 to 13, 38 to 43, and 59 to 71, respectively, and marked with asterisks) and a region involved in core homotypic interaction; domain II, which contains hydrophobic sequences for membrane association; and domain III, which serves as the signal sequence for E1, are also indicated (panel 1). pCAGGS-HACore-based mutant plasmids encoding substitutions in the signal peptide at the residues as indicated were also constructed (panel 2). Dashes indicate that the residue in that position of the mutant core protein is identical to that of the WT core. The Δ88-106 and Δ42-68 mutants, and their corresponding deleted regions are shown in panel 3. (B) 293T cells were transfected with the 191 core-expressing plasmid, and the calcium phosphate-DNA complexes were washed away 4 h posttransfection. Cells were then incubated at 37°C for different times as indicated and harvested, and the postnuclear fractions were subjected to Optiprep subcellular fractionation, followed by Western blot analysis with an HA MAb. The blots were scanned, and the percentages of core proteins distributed into soluble fractions 16 to 22 of the total core populations are indicated on the right of each panel. (C) The steady-state 191 core proteins localized in membrane-associated fractions 7 and 8 after the membrane flotation assay (data not shown) were combined, mixed with an equal volume of cold TE buffer, and then centrifuged in a Beckman SW41 rotor at 36,000 rpm for 2 h. The pelleted membranes were lysed with 0.5 ml of PBS containing 1% Triton X-100 (panel 1) or SDS (panel 2) at 37°C for 10 min prior to 5 to 50% sucrose gradient centrifugation. The combined cytosolic fractions 1 to 3 after the membrane flotation assay (data not shown) were diluted with 6 volumes of cold TE buffer and concentrated by Amicon Centricon plus-70 membranes (Millipore, Bedford, MA). The samples (0.5 ml) were treated with a final concentration of 1% Triton X-100 (panel 3) or SDS (panel 4) before 5 to 50% sucrose gradient centrifugation. The scheme illustrating these procedures is shown at the top.