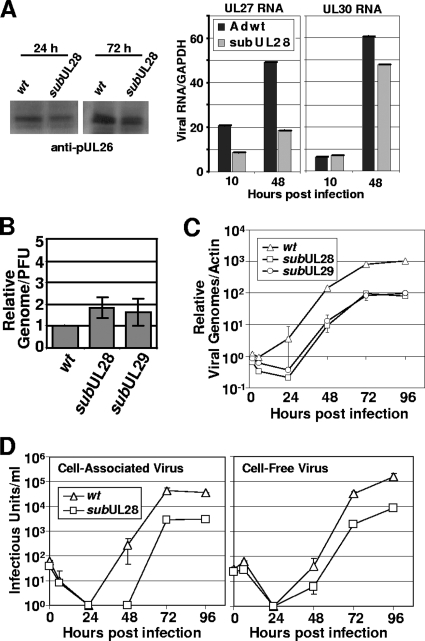
FIG. 4.
Inefficient BADsubUL28 and BADsubUL29 replication. (A) Expression of UL26, UL27, and UL30 in fibroblasts infected with subUL28 compared to wild-type virus. Fibroblasts were infected using equivalent amounts of virus, harvested at the indicated times, and processed for Western blot analysis with a pUL26-specific antibody. Expression of RNAs containing UL27 and UL30 ORFs were quantified by qRT-PCR using total RNA harvested from cells infected by wild-type and subUL28 viruses. (B) Infectivity of wild-type, subUL28, and subUL29 viruses. Genome content was determined by using qPCR and primers against HCMV DNA using partially purified virus, and infectious units were determined by quantifying IE1-positive cells using the same virus stock. The results are from duplicate experiments and are presented relative to wild-type virus. (C) Accumulation of viral DNA. Fibroblasts were infected with wild type, subUL28, or subUL29 at an input genome number equivalent to 0.1 infectious unit of wild-type virus/cell. Total cell-associated DNA was isolated, and viral genomes were quantified by using real-time PCR and normalized to β-actin DNA. (D) Single-step growth analysis of subUL28. Cell-associated (left panel) and cell-free (right panel) virus was assayed. Fibroblasts were infected with wild-type or subUL28 virus at an input genome number equivalent to 0.1 infectious unit of wild-type virus/cell. Infected cell culture medium was collected as cell-free virus samples, and cell-associated virus was isolated by freezing and thawing cell pellets. The amount of virus present in each sample was determined by counting the number of IE1-positive cells and experiment was completed in duplicate. wt, wild type.