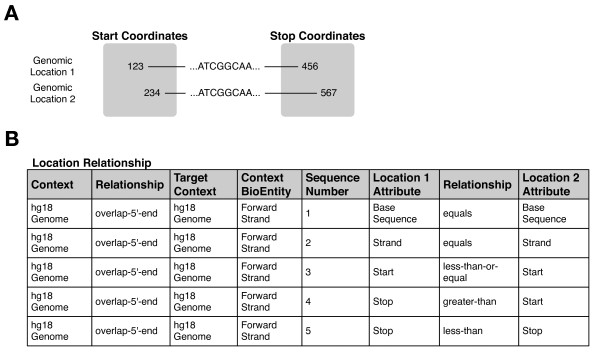
Figure 8.
Example ruleform for querying Locations by Relationship. Panel A: schematic of two locations from the 18th draft of the human genome declared in the system to have some overlap, with Genomic Location 1 overlapping the 5' end of Genomic Location 2. This relationship can be specified in terms of relationships between corresponding pairs of Attributes of those Locations. Panel B: We make the types of relationships we wish to query explicit, and can now allow us to perform queries based on Locations. For example, we define here a set of 5 rules that specify a relationship of "Location 1 overlaps the 5'-end of Location 2" that holds between Locations in the 18th draft of the human genome (specified by the "Context" and "Target Context" columns). Other ruleforms (not shown) contain meta-rules that ensure the appropriate rules are chosen for a given scenario. In this case, the rules for determining a 5' overlap depend on what nucleic acid strand (forward or reverse) a given genomic region lies; here we show the rules that apply for the forward strand only. These rules specify what relationships must hold between attributes of our query Location ("Location 1 Attribute" consideration) and those of any other Location ("Location 2 Attribute" consideration) whose 5' region is overlapped by our query Location. For instance, the first rule states that both Locations must be on the same sequence (i.e., chromosome), while the second states that they must be on the same strand. The remaining rules constrain relationships between the start and stop positions of the Locations such that the desired relationship holds. Any kind of relationship that can be expressed in terms of relationships between Location Attributes can be represented in this way.