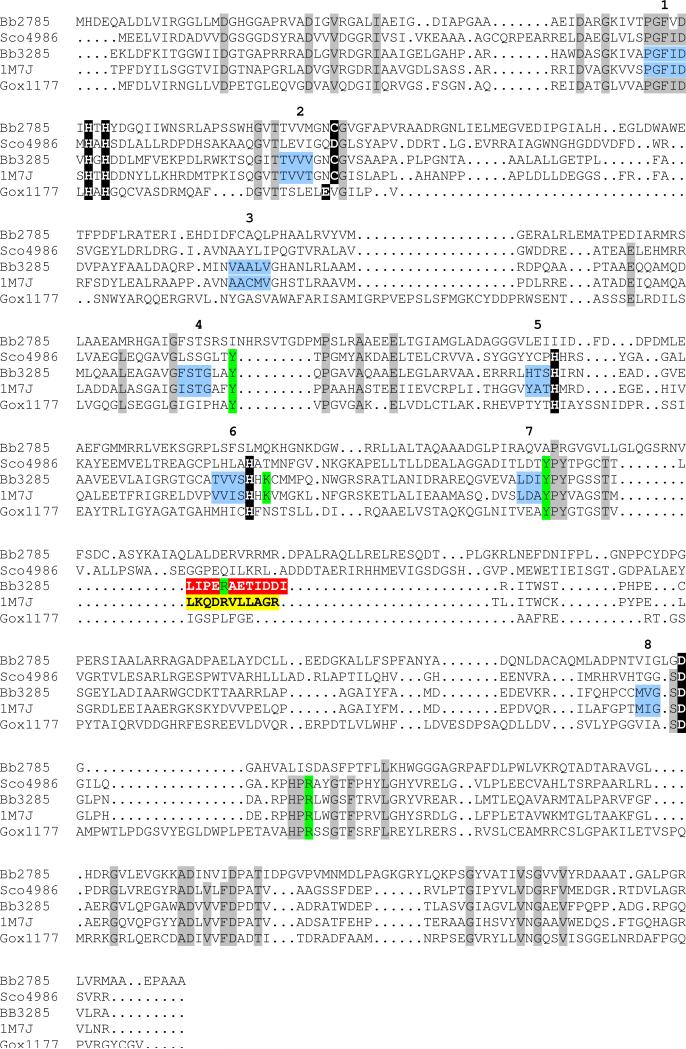
Figure 1.
Amino acid sequence alignment for the d-aminoacylase from A. facaelis (1M7J), Gox1177 from G. oxydans, Bb3285 and Bb2785 from B. bronchiseptica, and Sco4986 from S. coelicolor. Conservation patterns across these sequences with respect to the metal ligands identified in Bb3285 (see Figure 8) and the d-aminoacylase from A. facaelis are highlighted with a black background. The amino acid residues proposed to play a role in the recognition of the substrate in the active site of Bb3285 are highlighted in green. The variable substrate specificity loops in Bb3285 (291−302) and the DAA from A. facaelis (292−302) are highlighted in red and yellow, respectively. Those residues which represent the β-strands of the (β/α)8-barrel are colored light blue and the β-strands in the barrel are numbered. Those residues that are conserved in DAA (1m7j), Gox1177, Bb3285, and Sco4986 are highlighted with a grey background.