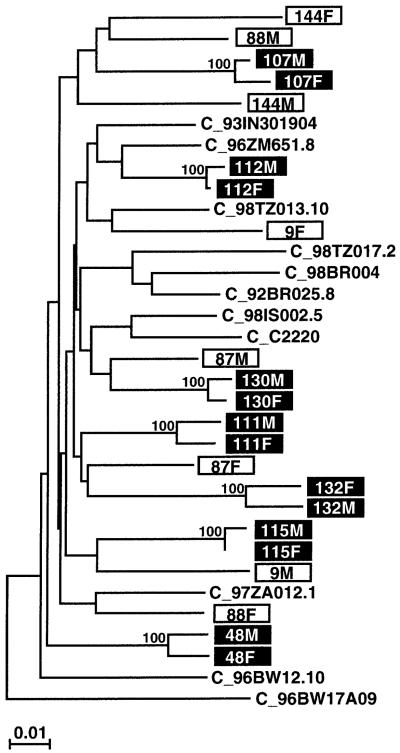
FIG. 1.
Epidemiological linkage analysis for a subset of putative HIV-1 transmission pairs. A phylogenetic tree was constructed from partial gp41 sequences (consensus length, 389 bp), using the neighbor-joining method25a and the Kimura two-parameter model.25b Horizontal branches are drawn to scale (the scale bar indicates 1% sequence divergence); vertical separation is for clarity only (the tree was rooted with A_U455). The bootstrap values at each node represent the percentage of 1000 bootstrap replicates that support the branching order (only values of 80% or higher are shown). Newly derived sequences from 22 different individuals (11 couples) are shown (boxed), along with 11 subtype C reference sequences (http://hiv-web.lanl.gov/HTML/alignments.html) from the Los Alamos sequence database (M and F indicate sequences derived from male and female partners, respectively). Viruses from seven couples (highlighted) are closely related to one other (0.5-2.7% nucleotide sequence diversity) and cluster together with significant bootstrap values (100%), thus indicating epidemiological linkage. By contrast, viruses from four other couples (boxed) do not cluster together and their range of within-couple diversity (5.9-10.1%) is comparable to that of the reference sequences (4.4-11.6%), thus making viral transmission between partners highly unlikely.