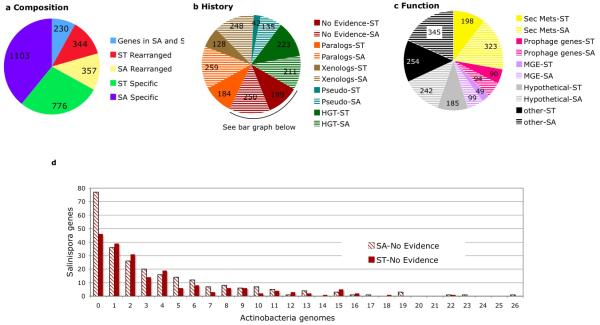
Figure 3.
Composition, evolutionary history, and function of island genes in S. tropica (ST) and S. arenicola (SA). (a) 3040 genes comprising 21 genomic islands were analyzed for positional orthology (ie., the gene is part of the shared “core” genome), re-arranged orthology (ie., the gene is present in the other genome but not in the same position or island), and species-specificity (gene totals presented in wedges). (b) The ST and SA species-specific island genes were analyzed for evidence of parology, xenology, and HGT. Pseudogenes and the number of genes with no evidence for any of these processes were also identified. (c) Functional annotation of the species-specific island genes. (d) Distribution of species-specific island genes that have no evidence for HGT or parology among 27 Actinobacterial genomes.