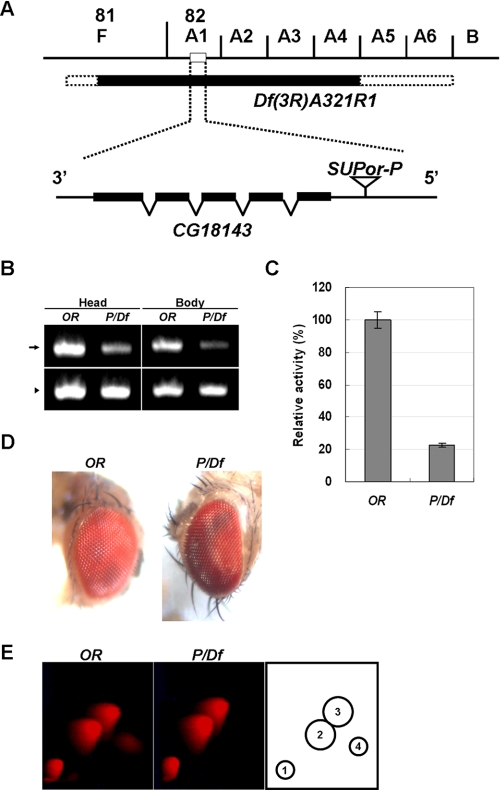
FIGURE 4.
Mutational analysis of the function of CG18143 in aurodrosopterin biosynthesis. A, cytological map of chromosome 3R showing CG18143 and its environs. The solid bar represents the deficiency region, including CG18143, in the Df(3R)A321R1 fly. Deficiency breakpoint limits are depicted by stippled bars. The SUPor-P element of the ;;P{y+mDint2 wBR.E.BR SUPor-P} fly is indicated 98 bp upstream from the first exon of CG18143. B, comparison of CG18143 mRNA level (arrow) between Oregon-R (OR) and SUPor-P/Df(3R)A321R1 (P/Df) transheterozygotes. Total RNA was extracted and reverse transcribed followed by PCR with specific primers as described under “Experimental Procedures.” The levels of glyceraldehyde-3-phosphate dehydrogenase mRNA (arrowhead) were used as internal controls. C, relative activities of dihydropterin deaminase in wild type flies (OR) and mutant flies (P/Df). The ammonium sulfate fraction (30–60%) from 2-day-old adult head extracts was used as the source of the enzyme. Vertical bars represent mean ± S.E. D, eye color phenotype of 2-day-old wild type and P/Df transheterozygote mutants. E, analysis of red pigments in the heads of Oregon-R (left panel) and P/Df transheterozygote (middle panel) by two-dimensional TLC. The fluorescent spots were detected by a long wave UV lamp. In P/Df transheterozygotes, aurodrosopterin was specifically decreased compared with the wild type flies. The identity of each spot is as follows: 1, neodrosopterin; 2, drosopterin; 3, isodrosopterin; and 4, aurodrosopterin (right panel).