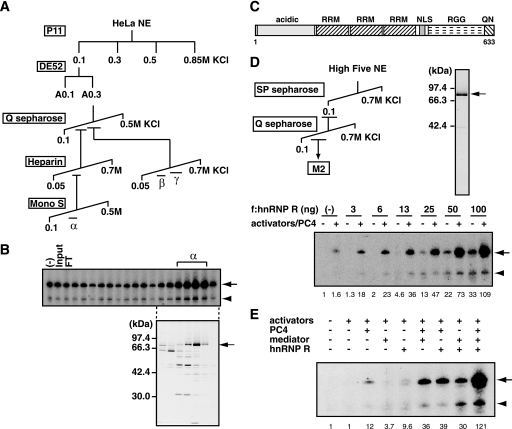
FIGURE 6.
hnRNP R stimulates transcription from the c-fos promoter. A, purification scheme of TREF activities. The TREF activity (A0.3) was further purified by Q Sepharose, heparin-Sepharose, and Mono S columns and separated into three chromatographically distinct activities designated TREFα, TREFβ, and TREFγ. B, transcriptional activities of the Mono S fractions of TREFα were assayed using pfMC2AT in the presence of SRF, Elk-1, CREB, ATF1, and PC4. The corresponding fractions were analyzed by SDS-PAGE and silver staining. The position of a ∼80-kDa protein that correlated with the transcriptional activity is indicated on the right. C, structural domains of hnRNP R. RRM, NLS, and RGG indicate the RNA recognition motif, nuclear localization signal, and arginine-glycine-glycine-rich domain, respectively. The regions rich in acidic amino acids or in glutamine and asparagine is indicated by acidic or QN, respectively. D, purification of recombinant hnRNP R. FLAG-tagged hnRNP R was expressed in insect cells and purified by SP Sepharose, Q Sepharose, and anti-FLAG M2-agarose. The purified hnRNP R was analyzed by SDS-PAGE. Transcriptional activity of hnRNP R was tested in the transcription assays using pfMC2AT, in which the presence of SRF, Elk-1, CREB, ATF1, and PC4 is indicated by +. Relative levels of activator-dependent transcription from pfMC2AT are indicated below the panel. E, hnRNP R, PC4, and Mediator were added in various combinations to the transcription reactions containing SRF, Elk-1, CREB, and ATF1 as activators. Relative levels of activator-dependent transcription from pfMC2AT are indicated below the panel.