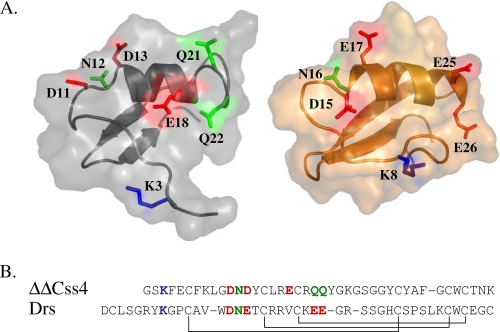
FIGURE 2.
Sequence alignment and three-dimensional structures of ΔΔCss4 and DRS. A, schematic diagrams of the Cα model structures of ΔΔCss4 and DRS covered by semitransparent molecular surfaces. The structure of DRS (right panel) is derived from the Protein Data Bank code 1MYN. The ΔΔCss4 model (left panel) is based on the NMR structure of Cn2 (Protein Data Bank code 1Cn2) and is spatially aligned with that of DRS. A was prepared using PyMOL. B, sequences were aligned according to the conserved cysteine residues, and the disulfide bonds formed between cysteine pairs are marked in solid lines. Dashes indicate gaps. Amino acid residues that were identified as part of the interacting surface of ΔΔCss4 with insect Navs (9) are shown in sticks according to their chemical nature (blue, positive charge; red, negative charge; green, nonpolar) and are also highlighted in the sequence alignment. Corresponding residues in DRS according to sequence and structural alignments are also shown in sticks.