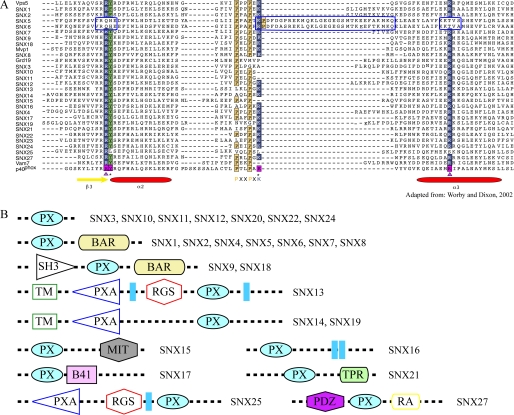
FIGURE 1.
Amino Acid sequence alignment of phox domains and domain architecture of the mammalian sorting nexin family. A, comparative sequence alignment of PX domains for residues equivalent to Gly49–Leu119 of the p40-PX domain (adapted from Worby and Dixon (21)). Prolines in the Pro-X-X-Pro motif are highlighted in yellow, and residues involved in phospholipid binding in the p40-PX domain are boxed in magenta. Arg58 and Arg105 are marked with magenta triangles, and Tyr59 and Lys92 are marked with black stars at the bottom of the sequences. The two conserved Arg residues and Lys92 of p40-PX in other PX domains are highlighted in dark blue boxes; those corresponding to Tyr59 are boxed in green. The secondary structure elements of p40-PX are indicated by yellow arrows (β-sheets) and red ovals (α-helices). The three sequence stretches that are unique in SNX5-PX (or SNX6-PX) are enclosed in a bright blue box. B, domain architecture of SNX family members. The four classes within the SNX family are designated as PX SNXs, PX-BAR SNXs, SH3-PX-BAR, and PX-other domain SNXs. Each individual domain is depicted in a different color and/or shape. The following domains are depicted: PX (phox), BAR (Bin-Amphiphysin-Rvs), SH3 (Src homology 3), TM (transmembrane), PXA (PX domain-associated), RGS (regulator of G-protein signaling), MIT (microtubule interacting and trafficking), B41 (band 4.1 homology), TPR (tetratricopeptide repeat), PDZ (postsynaptic protein PSD-95/SAP90, the Drosophila melanogaster septate junction protein Discs-large, and the tight junction protein ZO-1), and RA (Ras association).