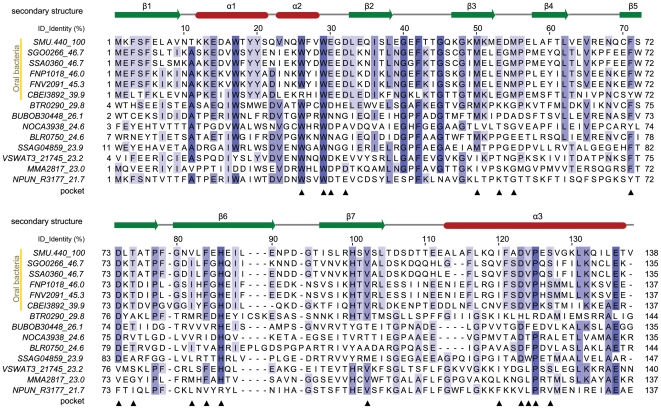
Figure 1. Structure-based multiple sequence alignment of SMU.440 homologs.
The corresponding NCBI RefSeq accession numbers and organisms are listed below, SMU.440, NP_720885, Streptococcus mutans UA159; SGO0266, YP_001449585, Streptococcus gordonii str. Challis substr. CH1; SSA0360, YP_001034364, Streptococcus sanguinis SK36; FNP1018, YP_002165260, Fusobacterium nucleatum subsp. polymorphum ATCC 10953; FNV2091, ZP_00143517, Fusobacterium nucleatum subsp. vincentii ATCC 49256; CBEI3892, YP_001310962, Clostridium beijerinckii NCIMB 8052; BTR0290, YP_001608752, Bartonella tribocorum CIP 105476; BUBOB30448, ZP_02382082, Burkholderia ubonensis Bu; NOCA3938, YP_925122, Nocardioides sp. JS614; BLR0750, NP_767390, Bradyrhizobium japonicum USDA 110; SSAG04859, YP_002179606, Streptomyces sp. Mg1; VSWAT3_21745, ZP_01813727, Vibrionales bacterium SWAT-3; MMA2817, YP_001354507, Janthinobacterium sp. Marseille; NPUN_R3177, YP_001866578, Nostoc punctiforme PCC 73102. The secondary structure was annotated based on the SMU.440 crystal structure. Residues highlighted with colored boxes are conserved to a varying extent, which is illustrated by the darkness of the color. For each protein, name and its sequence identity to SMU.440 are shown in the text columns to the left. Residues forming the cavity are marked by black triangles below the alignment.