Figure 1. Sequence analysis of HemG.
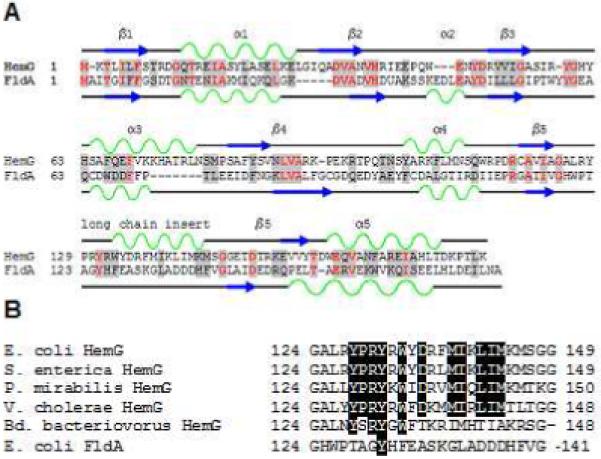
A. Sequence and secondary structure similarity of HemG with Flavodoxin A of E. coli. HemG is predicted by Jpred (33) to possess a typical flavodoxin-fold motif but with one notable difference, the presence of an alpha helical domain within the long chain insert loop. In the diagram similar residues are highlighted in grey, with conserved residues in red. Alpha helices and beta sheets are shown in green and blue respectively for each protein. B. Sequence alignment of the long chain insert loop of E. coli HemG with that of other annotated HemG proteins and FldA. Identical residues within the predicted alpha helix are highlighted. Significant change in this region (i. e. Bd. bacteriovorus) results in loss of function.
