Abstract
In order to speed up the process of cross-linked peptide identification and characterization, we have previously reported the development of Pro-CrossLink 1, a suite of software tools consisting of three programs, DetectShift, IdentifyXLink, and AssignXLink for mass spectrometric data analysis. Since its public disclosure, Pro-CrossLink has been downloaded by 101 research groups. Pro-CrossLink users have provided us with valuable feedback on the use of the program DetectShift. Here we assess some reasons for the generation of false positives by DetectShift. In addition, we provide users with suggestions on optimal parameter setting and efficient use of the software program.
INTRODUCTION
Chemical cross-linking in combination with mass spectrometry is a powerful tool to map protein structures and molecular interfaces in protein complexes.2-5 Among available strategies to identify cross-linked products, 18O-labeling via proteolysis is a useful tool. 6-9 However, analysis of the large set of mass spectrometric data generated by 18O-labeling experiments is challenging because of inherent complexity of cross-linking reaction mixtures. To address this issue, we developed DetectShift,1 a software program included in the software package Pro-CrossLink,1 that selects cross-linked peptide pair candidates incorporating more than two 18O atoms. For a set of mass spectrometric data containing one run for the 16O-labeled peptides and the other for the 18O-labeled peptides, DetectShift analyzes tens of thousands of mass spectrometric scans to calculate the charge states of the peptide ions, and then compute isotopic incorporation for the corresponding 16O-labeled and 18O-labeled ions within a user specified retention time window. Because of the complexity of the data set, false positives may be generated by DetectShift. In this manuscript, by describing major causes for the generation of false positives, we provide users with perspectives on DetectShift, and a guide to reduce the number of false positives.
EXPERIMENTAL
Cross-linking Reactions, Proteolytic Digestion and Mass Spectrometric Analysis
Cross-linking reactions, protein complex digestion and mass spectrometric analysis were performed as previously described.1
Development of Program DetectShift
DetectShift selects peptides incorporating a user-specified number of 18O atoms by analyzing multiply charged peptide ions obtained from ESI-MS. The original *.raw files acquired with the MassLynx software (Micromass, Cambridge, UK) are converted from profile data to centroid data using the MassLynx Accurate Mass Measure function and all the centroid data are exported into *.txt files using the MassLynx DataBridge function. Two types of signal intensity threshold can be specified: 1) an absolute intensity threshold for all signals in all scans and 2) a percentage value, which is multiplied by the base peak intensity in a scan to yield the intensity threshold for all signals in that scan. DetectShift first builds a list of all precursor ions in the 16O-digest based on their isotopic envelopes. The isotopic peaks of doubly, triply, quadruply and quintuply charged ions are separated by 0.5, 0.33, 0.25 and 0.2 m/z, respectively. If the number of a group of consecutive peaks exceeds a user-specified number “Nunlabeled”, these consecutive peaks are considered isotopic peaks of a precursor ion. Through the observation of a low abundant but recognizable peptide ion in an MS scan with low total ion current, “Nunlabeled” can be specified as the number of the isotopic peaks of the peptide ion that are above the user-specified signal threshold. The charge state of the precursor ion is determined by the distances between neighboring isotopic peaks. “Instrument Error” is user-specified which allows ± error variation for each isotopic peak within the isotopic envelope. Once the existence of a precursor ion is confirmed, the monoisotopic peak of this precursor ion is used to determine the extent of 18O atom incorporation by checking the existence of corresponding peaks in the 18O-digest. DetectShift is designed to search for peptides incorporating a user-specified number of 18O atoms. As an example, an incorporation of four 18O atoms causes a mass shift of 8.02 Da. For doubly, triply, quadruply and quintuply charged ions, the mass shifts are 4.01 m/z, 2.67 m/z, 2.00 m/z and 1.60 m/z, respectively. For a doubly charged ion, DetectShift searches for the existence of the peak in the 18O-digest whose value equals “the monoisotopic peak in the 16O-digest + 4.01 m/z ± error”. If the corresponding peak, representing the incorporation of four 18O atoms, exists in the 18O-digest, DetectShift continues to search for isotopic peaks following this peak. If the number of the isotopic peaks in the 18O-digest exceeds a user-specified number “Nlabeled”, DetectShift decides that the precursor ion is a cross-linked peptide pair candidate. The specification of the “Nlabeled” value follows the same rule as that of the “Nunlabeled” value. Because of the aforementioned common retention time shift problem that can occur between different chromatographic runs, even of the same sample, signals of the 16O-digest are compared to those of the 18O-digest within a certain retention time window, which can be specified by either minutes or MS scans.
RESULTS AND DISCUSSION
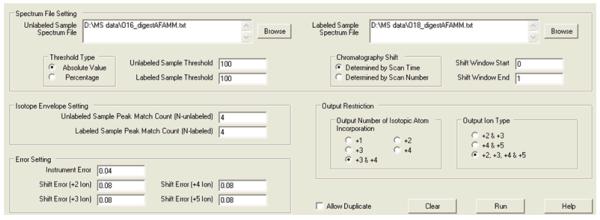
We have previously reported the use of the software program DetectShift to select cross-linked peptide candidates in the cytochrome P450 2E1 (CYP2E1) and cytochrome b5 (b5) complex.9 In that study, a CYP2E1-b5 complex was generated as a result of the treatment of an equimolar mixture of CYP2E1 and b5 with the cross-linking reagent 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC). 16O- and 18O-labeled tryptic digests of the complex were analyzed by nano-LC on an electrospray ionization quadrupole time-of-flight mass spectrometer (ESI-QTOF MS). Each LC-MS analysis contained ∼ 33,000 peptide ions. An additional complication was that the retention time of the ions in the 16O-digest lagged behind that of the corresponding ions in the 18O-digest by approximately 0.5 to 1 min because of the use of a flow splitter prior to the chromatographic column. As we reported, DetectShift narrowed the number of cross-linked peptide pair candidates from ∼ 33,000 precursor ions to 29 within minutes with the parameters provided (Figure 1). Visual inspection of the 29 DetectShift selected ions revealed that 14 were bona fide candidates and the other 15 false positives.
Figure 1.
The interface of DetectShift and the parameter settings for the selection of cross-linked peptide pair candidates.
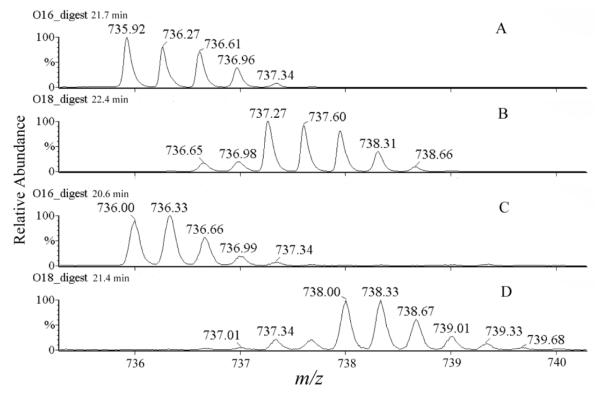
Through analyzing the results, we conclude that the major causes of false positive generation by DetectShift are: 1) Two precursor ions with the same charge state that have nearly the same mass elute within a narrow retention time window. For example, Figure 2A and Figure 2C show two such ions in the 16O-digest, at m/z 735.92 (21.7 min) and at m/z 736.00 (20.6 min). Both ions were selected by DetectShift as candidate ions, because an 18O-labeled peptide ion at m/z 738.00 (21.4 min) (Figure 2D) matched both ions shown in Figures 2A and 2C with incorporation of three 18O atoms. Subsequent manual inspection confirmed that the peptide ions in Figure 2C and Figure 2D were corresponding 16O-labeled and 18O-labeled ions, while the peptide ions in Figure 2A and Figure 2D were not. This is because most ions in the 16O-digest lagged behind the corresponding ions in the 18O-digest by 0.5 min to 1 min in the two chromatographic runs. The retention time shift (∼ 0.8 min) between the ions shown in Figures 2C and 2D agreed with such a systematic shift. However, the retention time shift (∼ -0.3 min) between the ions shown in Figures 2A and 2D did not. Further inspection assigned the ion in Figure 2A to another 18O-labeled peptide ion at m/z 737.27 (Figure 2B), with an incorporation of only two 18O atoms. Therefore, the ion in Figure 2C is a bona fide signal of a cross-linked peptide pair, and the ion in Figure 2A is a false positive selected by DetectShift.
Figure 2.
Single ion chromatograms of peptides from the 16O-digest (A & C) and the 18O-digest (B & D) in QTOF-ESI MS runs. [M + 3H]3+ = 735.92 at 21.7 min (A), [M + 3H]3+ = 737.27 at 22.4 min (B), [M + 3H]3+ = 736.00 at 20.6 min (C) and [M + 3H]3+ = 738.00 at 21.4 min (D).
Specifying a more strict retention time shift window based on the experimental results helps to decrease the number of such false positives. As shown in Table 1, the wider the retention time shift window, the smaller percentage of bona fide candidates in total results. The selection of a retention time shift window from -1 min to 1 min in this case produces about 39 results, among which 14 are bona fide candidates. Narrowing the retention time shift window from 0 min to 1 min increases the bona fide candidate percentage from 36% to 48% without losing any genuine result. However, narrowing the retention time shift window further from 0.5 min to 1 min causes the loss of two genuine candidates, even though the bona fide candidate percentage is enhanced. We notice that when running the program, a 5% to 15% widening of experimentally observed retention time shift window sometimes, but not always, helps to slightly increase the bona fide candidate percentage. However, widening the retention time shift window by more than 15% significantly increases false positive generation without notable improvement of bona fide candidate percentage.
Table 1.
Results obtained when different parameters are chosen to run the software program DetectShift.
| Parameter | Parameter Setting | Total Results | The Number of Bona Fide Candidates |
The Number of Lost Bona Fide Candidates |
The Pe rcentage of Bona Fide Candidates in Total Results |
|
|---|---|---|---|---|---|---|
| Retention Time Window | Shift Window Start (min) | -1 | 39 | 14 | 0 | 36% |
| Shift Window End (min) | 1 | |||||
| Shift Window Start (min) | 0 | 29 | 14 | 0 | 48% | |
| Shift Window End (min) | 1 | |||||
| Shift Window Start (min) | 0.5 | 23 | 12 | 2 | 52% | |
| Shift Window End (min) | 1 | |||||
| N Value (N-unlabeled = N-labeled) |
3 | 95 | 14 | 0 | 15% | |
| 4 | 29 | 14 | 0 | 48% | ||
| 5 | 8 | 8 | 6 | 100% | ||
| Intensity Threshold (Absolute Value) |
50 | 76 | 14 | 0 | 18% | |
| 100 | 29 | 14 | 0 | 48% | ||
| 200 | 10 | 9 | 5 | 90% | ||
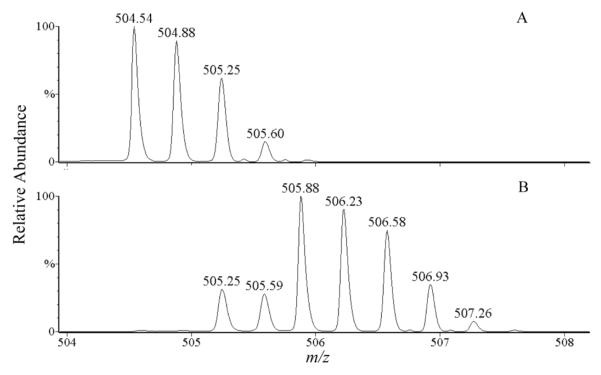
2) For a monoisotopic peak of high intensity, many subsequent isotopic peaks may exist with intensities above a specified threshold. An example is shown in Figure 3. For the triply charged ion at m/z 504.54 in the 16O-digest (Figure 3A), its corresponding 18O-labeled peptide ion at m/z 505.88 is shown in Figure 3B. The m/z shift of 1.34 for this triply charged ion suggests an incorporation of two 18O atoms in the peptide. However, in the 18O-digest (Figure 3B), one peak at m/z 506.58 in the isotopic envelope is followed by at least three isotopic peaks. DetectShift regarded the peak at m/z 506.58 as an extra monoisotopic peak corresponding to the monoisotopic peak at m/z 504.54 in the 16O-digest. The shift from m/z 504.54 to m/z 506.58 represents an incorporation of three 18O atoms for the triply charged ion. Thus, the peptide was selected by DetectShift as a cross-linked peptide pair candidate.
Figure 3.
Single ion chromatograms of peptides from the 16O-digest (A) and the 18O-digest (B) in QTOF-ESI MS runs. [M + 3H]3+ = 504.54 at 23.9 min (A) and [M + 3H]3+ = 505.88 at 24.4 min (B).
Increasing the required numbers of peak matches, “Nunlabeled” and “Nlabeled”, helps to reduce such false positives. It is shown in Table 1 that when “4” is set as the N value, the bona fide candidate percentage significantly increases, compared to when “3” is set as the N value (48% versus 15%). However, a further increase of the N value to “5” causes a loss of six genuine candidates among 14, even though a bona fide candidate percentage of 100% can be achieved. The selection of the N value depends on the quality of mass spectrometric data and the setting of intensity threshold. The higher the quality or the lower the threshold, the larger the N value should be set. We recommend that the set of N value start at “3” or “4”.
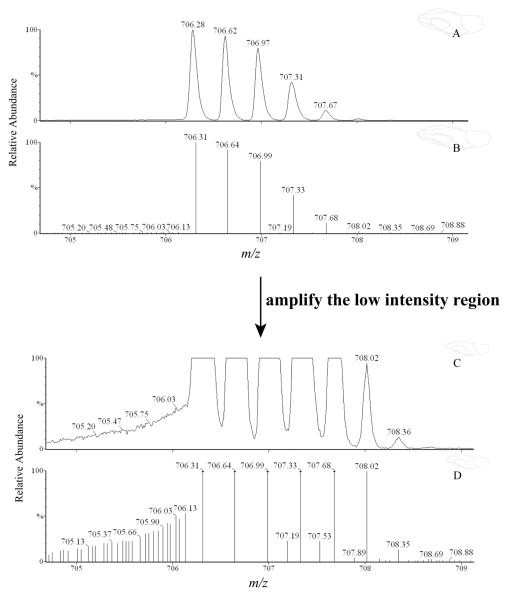
3) When centroid data are acquired, several peaks may appear around a major peak due to “shouldering” (Figure 4). When the intensity threshold is set lower than the intensities of these noise peaks, the inclusion of such peaks in the program calculation may lead to false positives. Selection of an optimal intensity threshold based on experimental results helps to reduce false positives without losing genuine results. Shown in Table 1, for the example data set, “100” is an optimal intensity threshold value, which yields a reasonable bona fide candidate percentage (48%) and a complete identification of bona fide candidates. Using “50” as the threshold generates a large set of results and thus a low percentage of bona fide candidates (18%), while using “200” causes a loss of 5 bona fide candidates.
Figure 4.
Single ion chromatogram of the peptide ion [M + 3H]3+ = 706.28 (A). Trace B shows the centroid signals of the signals shown in trace A. Trace C and D show the low intensity regions of trace A and B, respectively.
Although false positives are generated as discussed above, DetectShift dramatically simplifies the process of data analysis. Instead of manually sifting through ∼ 33,000 precursor ions, DetectShift selected 29 ions, which could be inspected visually to generate a list of 14 cross-linked peptide pair candidates.
It should be noted that although DetectShift was originally designed for selection of candidates of cross-linked peptide pairs, the use of 18O-labeling for other research purposes19 broadens the potential application of the program. In order to suit different applications, the number of 18O atoms incorporated into peptides is set as a user-specified parameter. In addition, charge states of output peptide ions are set as user-specified parameters because cross-linked peptide pairs, with their complex structures, generally exist in higher charge states than linear peptides under the same electrospray conditions.
CONCLUSION
In order to facilitate efficient use of DetectShift, we analyzed false positive results generated by the software program and provide insight into the optimal parameter settings. We hope the information provides a guide for DetectShift users. To access the program, please visit http://depts.washington.edu/ventures/UW_Technology/Express_Licenses/ProCrossLink.php
ACKNOWLEDGMENT
This work was supported by NIH grant GM32165 (SDN), the UW NIEHS sponsored Center for Ecogenetics and Environmental Health grant P30ES07033, an NCRR high end instrumentation award 1S10RR17262-01 (DRG) and the WWAMI RCE for biodefense and emerging infectious diseases 1U54AI57141-01 (DRG).
Footnotes
- CYP2E1
- cytochrome P450 2E1
- b5
- cytochrome b5
- EDC
- 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride
- ESI-QTOF MS
- electrospray ionization quadrupole time-of-flight mass spectrometry.
REFERENCES
- 1.Gao Q, Xue S, Doneanu CE, Shaffer SA, Goodlett DR, Nelson SD. Anal. Chem. 2006;78:2145. doi: 10.1021/ac051339c. [DOI] [PubMed] [Google Scholar]
- 2.Tang X, Munske GR, Siems WF, Bruce JE. Anal. Chem. 2005;77:311. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 3.Sinz A. J. Mass Spectrom. 2003;38:1225. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 4.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5802. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Back JW, de Jong L, Muijsers AO, de Koster CG. J. Mol. Biol. 2003;331:303. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 6.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Anal. Chem. 2002;74:4417. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 7.Collins CJ, Schilling B, Young M, Dollinger G, Guy RK. Bioorg. Med. Chem. Lett. 2003;13:4023. doi: 10.1016/j.bmcl.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 8.Huang BX, Kim HY, Dass C. J. Am. Soc. Mass Spectrom. 2004;15:1237. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Gao Q, Doneanu CE, Shaffer SA, Adman ET, Goodlett DR, Nelson SD. J Biol Chem. 2006;281:20404. doi: 10.1074/jbc.M601785200. [DOI] [PubMed] [Google Scholar]