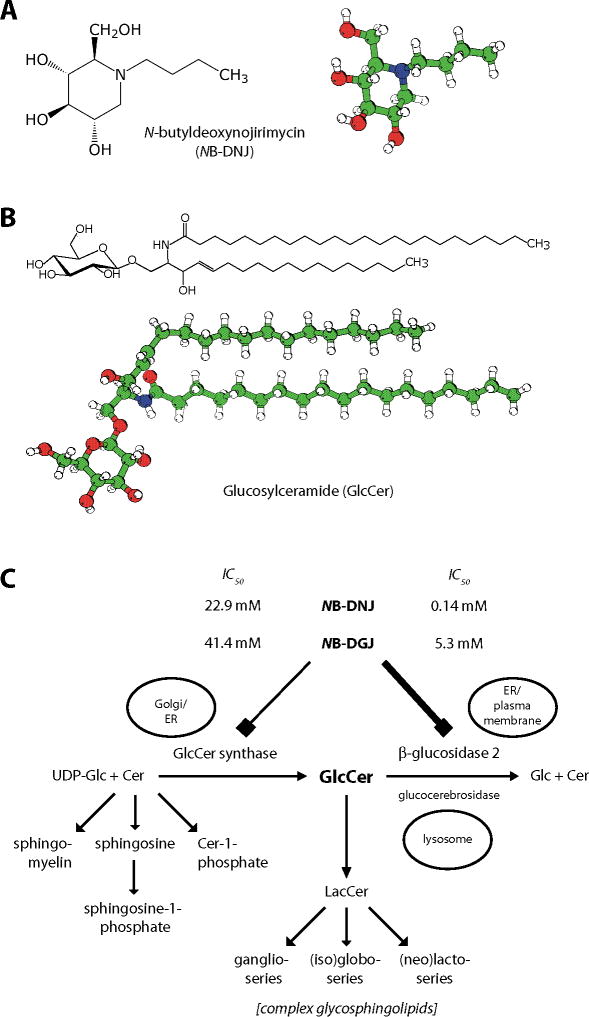
Figure 1. Structural formulas and metabolic pathways.
(A) Structural formula and model of NB-DNJ (miglustat, Zavesca®). The molecule differs from glucose in the presence of a nitrogen in the ring (where glucose has an oxygen), the butyl moiety linked to the ring nitrogen, and in the absence of a hydroxyl group from position 1. (B) Structural formula and model of glucosylceramide with an acyl chain of 24 carbons, without unsaturated bonds. (C) Metabolism of sphingolipids, focusing on glucosylceramide. Both the biosynthetic enzyme GCS as well as the degradative enzyme GBA2 can be inhibited by NB-DNJ and NB-DGJ, but GBA2 is much more sensitive to these compounds. This is evident from the differences between the IC50 values of the two drugs towards GCS and GBA2.
Colour code for molecular models: White: Hydrogen; Green: Carbon; Red: Oxygen; Blue: Nitrogen.
Cer: Ceramide; GBA2: β-glucosidase 2; GCS: GlcCer synthase; Glc: Glucose; GlcCer: Glucosylceramide; LacCer: Lactosylceramide; NB-DGJ: N-butyldeoxygalactonojirimycin; NB-DNJ: N-butyldeoxynojirimycin.
Panel (C) adapted from [6].