Abstract
Mutations in the rhodopsin gene that disrupt the encoded protein's folding properties are a major cause of autosomal dominant retinitis pigmentosa (ADRP). This disease is faithfully modeled in Drosophila where similar mutations in the ninaE gene, encoding rhodopsin-1 (Rh-1), cause ER stress and dominantly trigger age-related retinal degeneration. In addition, mutant flies bearing certain ninaE alleles have dramatically reduced Rh-1 protein levels, but the underlying mechanism for this reduction and significance of its contribution to the ADRP phenotype remains unclear. To address this question, we specifically analyzed the role of Drosophila genes homologous to the known yeast and animal regulators of the ER-associated degradation (ERAD) pathway, a process that reduces levels of misfolded proteins in the ER through proteasomal degradation. We found that loss-of-function of these putative ERAD factors resulted in increased levels of Rh-1 in ninaE mutant flies. Conversely, in an ER stress assay where mutant or wild-type Rh-1 were overexpressed in developing imaginal discs beyond the ER protein folding capacity of those cells, co-expression of certain ERAD factors was sufficient to reduce Rh-1 protein levels and to completely suppress ER stress reporter activation. Significantly, those ERAD factors that specifically reduced misfolded Rh-1 in the imaginal disc assay also delayed age-related retinal degeneration caused by an endogenous ninaE allele, indicating that ERAD acts as a protective mechanism against retinal degeneration in the Drosophila model for ADRP. These results suggest that manipulation of ERAD may serve as a powerful therapeutic strategy against a number of diseases associated with ER stress.
Keywords: apoptosis, endoplasmic reticulum, rhodopsin, unfolded protein response
The endoplasmic reticulum (ER) is an organelle in which membrane and secretory proteins are synthesized and folded into stable conformations. Reflecting the ER's essential role in protein folding, mutations that either cause misfolding of proteins synthesized in the ER or interfere with ER quality control mechanisms are frequent causes of degenerative diseases (1, 2).
Among the diseases associated with protein misfolding in the ER are class II autosomal dominant retinitis pigmentosa (ADRP), in which dominant rhodopsin mutations trigger age-related retinal degeneration and blindness (3, 4). The Drosophila genome has several rhodopsin genes, including ninaE that encodes the rhodopsin-1 protein (Rh-1) (5, 6). Exhibiting a striking similarity with human rhodopsin mutants, a number of ninaE alleles dominantly cause age-related retinal degeneration in Drosophila (7, 8). The amino acid substitutions that result from these ninaE mutations are similar to those human rhodopsin mutant proteins that fail to fold properly in cultured cells and underlie ADRP (8). In fact, Drosophila photoreceptors bearing these ninaE mutant alleles activate a specific transcriptional response that helps reduce misfolded proteins in this ER, widely referred to as the unfolded protein response (UPR) (9). Moreover, reducing the UPR through a mutation in a key component of this pathway, xbp1, aggravates the course of retinal degeneration in the Drosophila model (9). Similar observations have been made with the mammalian rhodopsin mutants (10), indicating that the pathology underlying ADRP is conserved between Drosophila and mammals.
Among a few unexplained features associated with these dominant ninaE alleles is a severe reduction of overall Rh-1 protein levels in the afflicted photoreceptors. While heterozygotes with a null ninaE allele have roughly half of the normal Rh-1 protein levels, many disease-causing rhodopsin alleles of humans and Drosophila lead to significantly lower rhodopsin levels under otherwise similar conditions (7, 8). However, whether such a reduction in Rh-1 levels has a functional significance in the retinal degeneration remains unclear. It is possible that the reduction of overall Rh-1 levels may have a beneficial consequence in the ADRP model, as it may protect against toxicity associated with aggregates in the ER. Alternatively, excessive Rh-1 reduction may accelerate retinal degeneration in ADRP, as insufficient levels of Rh-1 protein at the light-sensing compartment can compromise photoreceptor integrity and survival (11).
To understand the mechanism and function of Rh-1 reduction in this disease model, we investigated the role of putative regulators of Drosophila ER-associated degradation (ERAD). ERAD is regulated by a multiprotein complex that includes proteins involved in the recognition, retrotranslocation, and ubiquitination of misfolded proteins in the ER (12). Studies using Saccharomyces cerevisiae have suggested the existence of three major ERAD subpathways, defined by the subcellular location of the lesion that causes protein misfolding (13–15). A protein that misfolds in the ER lumen is thought to be degraded through the ERAD-L pathway. On the other hand, proteins with lesions in transmembrane domains are thought to be substrates of the ERAD-M pathway. Proteins with lesions on the cytoplasmic side of the ER are processed by the ERAD-C pathway. In yeast, each subpathway requires a distinct set of ERAD complex subunits. For example, the two yeast mannosidases that recognize misfolded glycoproteins, Htm1p, and membrane protein complex containing Usa1p and Derlin, appear dedicated to the ERAD-L pathway, since ERAD-M substrates bypass the requirement of those subunits (13–16). Whether ERAD regulation in metazoans follows the same rules remains unknown.
Using genetic tools of Drosophila, here we demonstrate that ERAD regulators help reduce stress and retinal degeneration caused by mutant Rh-1 in the Drosophila ADRP model and serve as a protective mechanism. Specifically, we show that disruption of the ERAD pathway leads to an increase in Rh-1 protein levels in the Drosophila model for ADRP. Conversely, overexpression of certain subunits of the ERAD machinery was sufficient to reduce the levels of ER stress-causing Rh-1 proteins. Individual subunits showed distinct specificity toward their substrates that were sometimes inconsistent with what is expected from the ERAD subpathways defined in yeast. Most significantly, certain factors were able to suppress late-onset retinal degeneration in a Drosophila model for ADRP. These results suggest that specific ERAD mechanisms can be exploited as a therapeutic strategy against conformational diseases.
Results
Loss of ERAD Components Partially Restores the Level of Rhodopsin-1 in the ninaEG69D−/+ Retina.
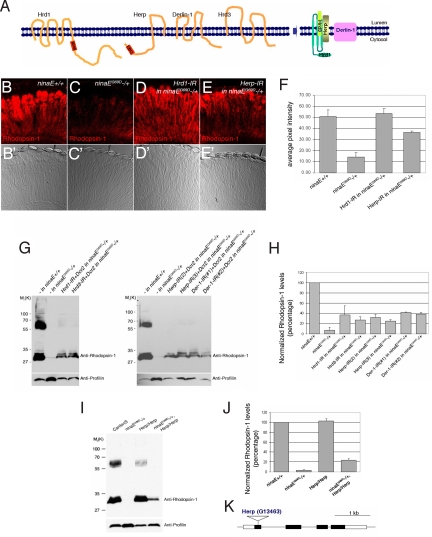
To study the mechanism and consequences of Rh-1 reduction in the Drosophila model for ADRP, we examined heterozygous flies bearing a ninaE allele with a Gly residue in a transmembrane domain replaced by Asp (henceforth referred to as ninaEG69D). Similar to human autosomal dominant retinitis pigmentosa (ADRP), these flies activate the UPR in their photoreceptors, which eventually undergo age-related retinal degeneration (7, 8, 11). To determine the role of ERAD in the pathogenesis of ADRP, we modulated the levels of Drosophila homologs of yeast and human ERAD regulator proteins, with a particular focus on those integral membrane proteins that interact with each other (12). One of these Drosophila gene products, known as septin interacting protein 3, was previously identified in a yeast two-hybrid assay (17). We noticed that this Drosophila gene is most homologous to yeast and human Hrd1, a ubiquitin ligase of the ERAD pathway (18–20), and we will henceforth refer to it as Hrd1. As in humans, Drosophila Hrd1 has six predicted transmembrane domains and a RING domain on the cytoplasmic side (Fig. 1A). Other genes we examined include a candidate retrotranslocation channel component, Derlin-1 (CG10908), and its associated proteins Hrd3 (CG10221) and Herp (CG14536) (Fig. 1A). When the retina of heterozygotic flies bearing the ninaEG69D allele were immunolabeled with anti-Rh-1 antibody, we found that the average anti-Rh-1 labeling intensity was reduced to approximately 28% of wild-type levels (Fig. 1C), consistent with previous reports (7, 8, 21, 22) that have documented the reduction of both wild-type and mutant Rh-1 proteins under these conditions. To determine whether this reduction is due, at least in part, to enhanced ERAD, we knocked down Hrd1 and Herp in the retina of ninaEG69D−/+ flies using inverted repeat transgenic constructs that are designed to produce double stranded (ds) RNA (see Materials and Methods). Knock down of Drosophila Hrd1 in this background restored the overall Rh-1 levels to an extent that was statistically indistinguishable from that of wild-type flies (Fig. 1D; n = 3, P = 0.6). Although to a lesser degree, Herp knock down also restored Rh-1 levels to a significant degree (Fig. 1E; n = 3, P = 0.0009). To complement these immunohistochemical analysis, we examined Rh-1 levels through Western blots under similar genetic conditions. The total amount of Rh-1 in ninaEG69D−/+ flies estimated through Western blots was 7% of the wild-type levels, which was lower than that assessed through immunohistochemistry. This is most likely because insoluble membrane proteins are frequently lost during SDS/PAGE sample preparation. However, the changes of Rh-1 levels in response to ERAD inhibition was consistent with that observed through immunohistochemistry, with the knock down of Hrd1 or other ERAD regulators partially restoring the Rh-1 levels in ninaEG69D−/+ flies (Fig. 1 G and H). The results were further validated using a Herp mutant allele, HerpG13463, which has an EP- element inserted within its protein coding sequence (Fig. 1K). While the loss-of Herp did not affect the total Rh-1 protein level in the ninaE+ background, it partially restored total Rh-1 levels in the ninaEG69D−/+ background (p = 0.0003) (Fig. 1 I and J). There was no statistically significant difference in the steady state Rh-1 levels between the Herp knock down and HerpG13463−/− flies (P = 0.45 and 0.1 for comparisons with two independent inverted repeat lines). While these results can be most simply interpreted as evidence of misfolded Rh-1 proteins being degraded by ERAD, an alternative scenario is also possible where defective ERAD may increase Rh-1 levels by inducing ER chaperons and enhance Rh-1 folding. In fact, blocking ERAD in yeast is known to stimulate UPR signaling and induce ER chaperones (23). However, such an indirect model appears unlikely in the Drosophila retina, as we find that the Herp mutant flies did not induce the expression of heat shock cognate protein 3 (hsc3), a major ER chaperon that is homologous to BiP and a well established target of UPR (Fig. S1). These observations support the idea that the examined Drosophila ERAD factors stimulate the degradation of misfolded Rh-1 in the ADRP model.
Fig. 1.
Loss of ERAD regulators partially restores the level of Rh-1 in the retina of ninaEG69D−/+ flies. (A) Schematic diagrams of the predicted membrane topology and domain structure of ERAD components through EMBnet-CH. (B–E) Horizontal retina cryosections stained with anti-Rh-1 antibody in 1- to 3-day-old flies. (B) A control ninaE +/+ retina. ninaEG69D−/+ retina (C) show low Rh-1 labeling, while Rh-1 levels recover when Hrd1 (D) or Herp (E) are knocked down in this genetic background. In all RNAi experiments, dicer2 (drc2) were co-expressed to enhance the efficiency of RNAi knockdown. (F) The average intensity of anti-Rh-1 labeling (n = 3), as quantified through the Image J. Error bars, ± SEM. (G and I) A Western blot of Drosophila adult head extracts of the indicated genotypes probed for Rh-1 (upper gel) and anti-profilin (as a loading control; lower gel). (H and J) Quantification of the average normalized Rh-1 band intensity as shown in (G and I) (average of n = 3; Der-1, average of n = 2), with the value from wild-type of Rh-1+/+ heads extracts set at 100%. (K) The structure of the Herp genomic locus. In the HerpG13463, an EP-element is inserted within its protein-coding region. Genotypes: Rh1-Gal4;;ninaE+/+ (B and B′), Rh1-Gal4;;ninaEG69D/+ (C and C′), Rh1-Gal4;UAS-Dcr2/+;UAS-Hrd1-IR/ninaEG69D (D and D′), Rh1-Gal4;UAS-Dcr2/+;UAS-Herp-IR/ninaEG69D (E and E′), (G, lanes 1 and 5) Rh1-Gal4;;ninaE+/+, (G, lanes 2 and 6) Rh1-Gal4;;ninaEG69D/+, (G, lane 3) Rh1-Gal4;UAS-Dcr2/+;UAS-Hrd1-IR/ninaEG69D, (G, lane 4) Rh1-Gal4;UAS-Hrd3-IR/+;UAS-Dcr2/ninaEG69D, (G, lane 7) Rh1-Gal4;UAS-Herp-IR/+;UAS-Dcr2/ninaEG69D, (G, lane 8) Rh1-Gal4;UAS-Dcr2/+;UAS-Herp-IR/ninaEG69D, (G, lane 9) Rh1-Gal4;UAS-Dcr2/+;UAS-Der-1-IR(#1)/ninaEG69D, (G, lane 10) Rh1-Gal4;UAS-Dcr2/+;UAS-Der-1-IR(#2)/ninaEG69D, (I, lane 1) CantonS, (I, lane 2) ninaEG69D/+, (I, lane 3) HerpG13463/HerpG13463, (I, lane 4) HerpG13463/HerpG13463;ninaEG69D/+.
ER Stress Caused by Rh-1 Misexpression Is Strongly Suppressed by Drosophila Hrd1.
As the retina of ninaEG69D−/+ flies contains a mixture of mutant and wild-type Rh-1 proteins that are similar in size, it is difficult to distinguish their individual fates through Western blots or immunohistochemistry. To examine the behavior of particular Rh-1 alleles, we expressed mutant or wild-type Rh-1 in the larval eye imaginal discs, a developing tissue that does not yet express Rh-1. To assess the level of ER stress caused by such Rh-1 misexpression, we used xbp1-EGFP as a reporter designed to detect the unconventional mRNA splicing of xbp1, which is a key signaling event for the initiation of UPR. This reporter allows EGFP to be expressed in frame, only when a 23-nt intron in xbp1 is spliced out by the ER stress-activated endonuclease ire-1, thereby marking with green fluorescence those cells suffering significant ER stress (9). While this marker is not activated in response to proteins that misfold in the cytoplasm (9), xbp1-EGFP fluorescence was detected in all examined eye discs in which Rh-1G69D or Rh-1WT were expressed (Fig. 2 F and H, n > 10). The observation that Rh-1WT also activates the UPR in these cells was unexpected, and may be due to the limited ER capacity of the imaginal disc cells to fold and export high levels of Rh-1.
Fig. 2.
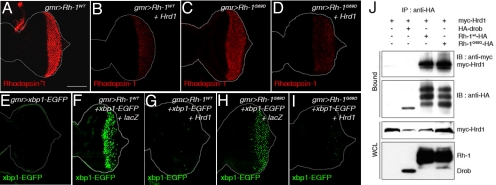
ER stress caused by Rh-1 misexpression is strongly suppressed by Drosophila Hrd1. (A–D) Representative images of eye imaginal discs expressing Rh-1WT (A) or Rh-1G69D (C) alone, or together with Hrd1 (B and D). Anti-Rh-1 antibody labeling is in red. E–I Hrd1 co-expression abolished ER stress caused by wild-type or mutant Rh-1 misexpression, as determined by the ER stress marker, xbp1-EGFP (green). Shown are representative discs expressing xbp1-EGFP alone (E), or together with indicated genes. (J) Co-immunoprecipitation assays between Hrd1 and Rh-1 in 293T cells. Hrd1 was tagged with the myc epitope, while Rh-1 was tagged with HA. HA-Drob-1 is a membrane protein used as a negative control. [Scale bars, 100 μm (A).] Genotypes: gmr-Gal4/+;UAS-Rh-1WT/+ (A), gmr-Gal4/UAS-Hrd1;UAS-Rh-1WT/+ (B), gmr-Gal4/+;UAS-Rh-1G69D/+ (C), gmr-Gal4/UAS-Hrd1;UAS-Rh-1G69D/+ (D), gmr-Gal4/+;UAS-xbp1-EGFP/+ (E), gmr-Gal4/UAS-lacZ;UAS-Rh-1WT/UAS-xbp1-EGFP (F), gmr-Gal4/UAS-Hrd1;UAS-Rh-1WT/UAS-xbp1-EGFP (G), gmr-Gal4/UAS-lacZ;UAS- Rh-1G69D/UAS-xbp1-EGFP (H), and gmr-Gal4/UAS-Hrd1;UAS-Rh-1G69D/UAS-xbp1-EGFP (I).
The establishment of the above assay allowed us to ask whether overexpression of any of the putative ERAD regulator genes could suppress the ER stress caused by Rh-1 misexpression. We found that this was the case, with Drosophila Hrd1 being one the strongest suppressors among those examined. In larval eye discs, Drosophila Hrd1 lowered Rh-1WT and Rh-1G69D protein levels when these were co-expressed through the eye-specific gmr-Gal4 driver (Fig. 2 A–D). The effect of Hrd1 on the misexpressed Rh-1 proteins was further validated through an alternative gene expression system that employs tubulin-Gal4 flip-out technology, which generates mosaic clones expressing genes of choice through the tubulin promoter (see Materials and Methods). When mosaic clones expressing Drosophila Hrd1 were generated within the population of Rh-1G69D-expressing cells, those clones had distinctively lower levels of Rh-1G69D protein, compared to the neighboring cells that did not express Hrd1 (Fig. S2 B and B′; n = 8). Hrd1 appeared to lower mutant and wild-type Rh-1 to levels that virtually eliminated ER stress itself, as evidenced by a near complete suppression of the xbp1-EGFP marker, which was otherwise activated in response to the misexpression in either wild type or mutant Rh-1 in eye discs (Fig. 2 E–I; n > 10). As an indirect measure of stress, we also examined markers for apoptosis, an active cell death program that involves, among others, proteolytic cleavage and activation of caspases. Labeling of imaginal discs with an antibody that detects such caspase cleavage event (24) showed that Drosophila Hrd1 co-expression suppressed the excessive apoptosis associated with Rh-1WT misexpression (Fig. S2 C and D; n = 8). To determine whether Hrd1 interacts with either Rh-1WT or Rh-1G69D, we performed co-immunoprecipitation assays. Myc-tagged Hrd1 was coexpressed in 293T cells along with HA-tagged Rh-1WT, Rh-1G69D or a control membrane protein, Drob-1 (25). Hrd1 co-precipitated with Rh-1WTand Rh-1G69D, but not with Drob-1 (Fig. 2J), establishing that Hrd1 forms a physical complex with mutant and wild-type Rh-1 and strongly suppresses ER stress caused by these Rh-1 proteins.
Overexpression of Putative ERAD Components in Drosophila.
The findings just described prompted us to test other Drosophila homologs of the ER associated degradation (ERAD) factors, including Hrd3, Herp, Derlin-1, and Derlin-2 (CG14899). In addition, we characterized the role of Drosophila EDEM homologs, CG3810 and CG5682, which we refer to as EDEM1 and EDEM2, respectively. Specifically, we examined their ability to suppress the rough eye phenotype generated by overexpression of Rh-1WT in larval eye discs, a condition that causes ER stress (Fig. S2). Consistent with the results obtained with the xbp1-EGFP assay, Hrd1 overexpression in eye imaginal discs almost completely rescued the rough eye phenotype in adults. Herp overexpression also suppressed the eye phenotype, but to a significantly lesser extent. On the other hand, other examined genes failed to rescue the partial eye ablation caused by Rh-1WT under similar conditions (Fig. S2).
Drosophila EDEM2 Overexpression Reduces Mutant, but Not Wild-Type Rh-1 Levels.
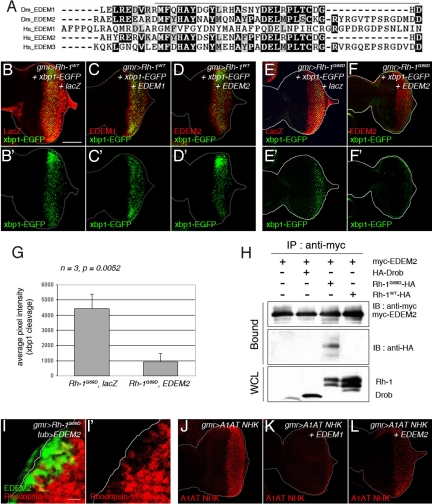
Although the above study suggested that the Drosophila EDEMs do not affect Rh-1WT levels, we further tested whether EDEMs affect other ERAD substrates. The Drosophila EDEM1 and EDEM2 are most homologous to mammalian EDEM2 and EDEM3 respectively (Fig. 3A) and also to the yeast Htm1p, which are best characterized as mannosidases that extract primarily luminal misfolded glycoproteins from their chaperone cycles and deliver them to the ERAD machinery (16, 26–28). Inconsistent with the idea that Htm1p is only dedicated to ERAD-L, overexpression of EDEM2 was able to reduce the levels of a likely ERAD-M substrate, Rh-1G69D, which has a mutation in a transmembrane domain (Fig. 3 I and I′; n = 6). More intriguingly, EDEM2 did not suppress ER stress caused by Rh-1WT, as assessed through xbp1-EGFP fluorescence (Fig. 3 B–D). The reduction of Rh-1G69D levels by EDEM2 was accompanied by a strong suppression of ER stress, as measured by xbp1-EGFP fluorescence (Fig. 3 E′, F′, and G; n = 3, P = 0.0052). EDEM1 overexpression neither affected Rh-1WT nor Rh-1G69D (Fig. 3C and Fig. S3D). We then performed immunoprecipitation assays to test whether EDEM2 forms a physical complex with Rh-1G69D or Rh-1WT. Consistent with EDEM2's effects on mutant and wild-type Rh-1 levels in vivo, EDEM2 co-precipitated with Rh-1G69D (Fig. 3H, lane 3) but not with Rh-1WT (Fig. 3H, lane 4). These results were further validated by the examination of the adult eye ablation phonotype. Similar to the case of overexpressing Rh-1WT, the misexpression of Rh-1G69D in larval eye discs generated partially ablated adult eyes (Fig. S3B). Although the degree of suppression was less than that observed with the Rh-1WT misexpressing eyes, Hrd1 scored as one of the strongest suppressors the Rh-1G69D overexpression phenotype, reducing the extent of eye tissue loss and de-pigmentation (Fig. S3C). Consistent with the results obtained with xbp1-EGFP in eye imaginal discs, EDEM2 overexpression suppressed the Rh-1G69D overexpression phenotype (Fig. S3E).
Fig. 3.
Drosophila EDEM2 overexpression reduces mutant, but not wild-type Rh-1 levels. (A) The amino acid sequence alignment between EDEM family proteins of Drosophila and humans, generated with the ClustalW algorithm. Dark shading indicates identity, whereas light shading indicates similarity. Solid lines indicate the catalytic domain of class I mannosidase (Glycosyl hydrolase family 47). The sequences shown, with their corresponding NCBI database accession numbers, are as follows: D. melanogaster (Dm) EDEM1 (NP_726777); D. melanogaster (Dm) EDEM2 (AAF53255); human (Hs) EDEM1 (AAH19088); human (Hs) EDEM2 (NP_060687); human (Hs) EDEM3 (NP_079467). (B–D) The effect of Drosophila EDEM1 and EDEM2 on misexpressed Rh-1WT. Shown are representative discs expressing Rh-1WT, together with lacZ (B), EDEM1 (C), or EDEM2 (D), or expressing Rh-1G69D with lacZ (E) or EDEM2 (F). The degree of ER stress is assessed through xbp1-EGFP activation (B′–F′, green). (G) Quantification of the xbp1-EGFP activation levels as shown in (E and F). Error bars, ± SEM. (H) EDEM2 physically interacts with Rh-1G69D in S2 cells. EDEM2 was immunoprecipitated through its myc-tag, and its interaction partners were detected through their HA-epitopes. (I and I′) A disc expressing EDEM2 (green) in a flip-out mosaic clone shows reduced levels of Rh-1G69D (red). (J–L) Both of EDEM1 and EDEM2 effectively downregulated the level of alpha 1-antitrypsinNHK (A1AT NHK, in red). (Scale bars, 100 μm (B) and 10 μm (I).) Genotypes: gmr-Gal4/UAS-lacZ;UAS-Rh-1WT/UAS-xbp1-EGFP (B and B′), gmr-Gal4/+;UAS-Rh-1WT, UAS-EDEM1/UAS-xbp1-EGFP (C and C′), gmr-Gal4/UAS-EDEM2;UAS-Rh-1WT/UAS-xbp1-EGFP (D and D′), gmr-Gal4/UAS-lacZ;UAS-Rh-1G69D/UAS-xbp1-EGFP (E and E′), gmr-Gal4/UAS-EDEM2;UAS-Rh-1G69D/UAS-xbp1-EGFP (F and F′), hs-flp;UAS-EDEM2/+;tub>GFP>Gal4/gmr-Rh-1G69D (I and I′), gmr-Gal4/UAS-alpha1-antitrypsinNHK;+/+ (J), gmr-Gal4/UAS-alpha1-antitrypsinNHK;UAS-EDEM1/+ (K), and gmr-Gal4/UAS-alpha1-antitrypsinNHK, UAS-EDEM2;+/+ (L).
In addition to the examination of Rh-1 levels, we also determined the effect of the EDEMs on an ER luminal ERAD substrate, alpha 1-antitrypsinNHK, which in mammalian cells, has been shown to cause ER stress that can be relieved by overexpression of mammalian EDEM2 or EDEM3 (27, 29). As expected from these studies, overexpression of Drosophila EDEM1 or EDEM2 reduced the levels of alpha 1-antitrypsinNHK (Fig. 3 J–L), which however was not affected by Hrd1 (Fig. S4). Collectively, these observations show that ERAD factors show specificity toward their substrates, but such specificity is not exclusively governed by the subcellular location of the lesion, as had been suggested previously (13–16).
Drosophila Hrd1 and EDEM2 Overexpression Suppresses Late Onset Retinal Degeneration in the Drosophila ADRP Model.
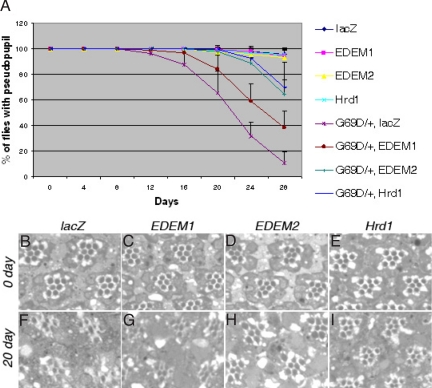
To determine the physiological significance of the ERAD specificity observed in the imaginal disc overexpression assay, we turned to the endogenous ninaEG69D mutant allele that dominantly causes age-related retinal degeneration. Specifically, we overexpressed Drosophila Hrd1 or EDEMs in the retina of ninaEG69D−/+ flies and examined their effects on the time course of retinal degeneration. To test this, we first used the pseudopupil assay, a noninvasive technique used to assess the regularity of the GFP-labeled photoreceptor array, which collectively projects to generate a single trapezoidal pseudoimage (30). While ninaE+/+ animals overexpressing only lacZ, EDEM1, EDEM2, or Hrd1 did not show any signs of retinal degeneration using this assay (Fig. 4A), ninaEG69D−/+ flies expressing a control protein, lacZ, had their pseudopupils disappear beginning at 12 days after eclosion, in a progressive manner, with only 10% of these flies showing intact pseudopupils at 28 days after eclosion (Fig. 4A; 10.47 ± 8.46%, n = 4). Significantly, the expression of Drosophila Hrd1 in a ninaEG69D−/+ background dramatically decreased the rate of deep pseudopupil loss with 70% of the examined flies showing intact pseudopupils at day 28 (Fig. 4A; 69.72 ± 19.58%, n = 4, P = 0.0014). The overexpression of EDEM2 in the ninaEG69D−/+ retina also delayed the time course of retinal degeneration significantly, with 64% of flies still showing intact pseudopupils at day 28 (Fig. 4A; 64.43 ± 11.11%, n = 4, P = 0.0002). Consistent with what we had observed in larval imaginal disc misexpression assays, EDEM1 had only a minimal effect on delaying retinal degeneration of ninaEG69D−/+ flies (Fig. 4A; 38.02 ± 13.47%, n = 4, P = 0.013).
Fig. 4.
Drosophila Hrd1 and EDEM2 overexpression suppresses late onset retinal degeneration of ninaEG69D−/+ flies. (A) Quantification of the extent of retinal degeneration through the pseudopupil assay. For each genenotype, the graph shows the percentage of flies with intact pseudopupils (average of four independent crosses). Specifically, lacZ, EDEM1, EDEM2 or Hrd1 were overexpressed through the Rh1-Gal4 driver in ninaE mutant or wild type backgrounds. Overexpression of Hrd1 and EDEM2 delay the course of retinal degeneration of ninaEG69D−/+ flies. (B–I) Representative images of adult retina overexpressing designated ERAD factors in the ninaEG69D−/+ background. (B–E) At day 0 after eclosion, retina of all genotypes have regular array of ommatidia. (F–I) Retinal sections of indicated genotypes at day 20. The degree of ommatidial disarray is suppressed by overexpressing EDEM2 or Hrd1 (H and I), but not by lacZ (F) or EDEM1 (G).
The pseudopupil results were independently validated by semithin sections of retinas of various genotypes. A single ommatidium of wild-type flies contains seven rhabdomeres, light-sensing organelles equivalent to the outer segments of mammalian rod cells (31). In 1-day-old ninaEG69D−/+ flies (i.e., 0 day light incubation) expressing lacZ, EDEM1, EDEM2, or Hrd1, retinas maintained a regular repeating pattern of ommatidia, with virtually all ommatidia containing seven rhabdomeres in a regular array (Fig. 4 B–E). In contrast, the 21-day-old ninaEG69D−/+ flies expressing a control lacZ gene had most ommatidia in disarray with large vacuoles and an overall reduction of rhabdomere numbers (Fig. 4F). Only a small percentage of the examined ommatidia retained all seven rhabdomeres within a unit (Fig. 4J; 2 ± 2%, n = 3). When EDEM2 or Hrd1 was overexpressed in the ninaEG69D−/+ retina under otherwise similar conditions, the degree of ommatidial disarray was significantly suppressed (Fig. 4 H and I, respectively). Specifically, a majority of the ommatidia expressing EDEM2 or Hrd1 retained all seven rabdomeres associated within a unit (Fig. 4 H and I; 68.9 ± 16.1%, n = 3, P = 0.002; 68.4 ± 6.7%, n = 3, P < 0.0001, respectively). Consistent with the partial suppression of pseudo pupil loss by EDEM1, ommatidial arrays in EDEM1 overexpressing ninaEG69D−/+ flies were only mildly restored (Fig. 4G; 27.8 ± 4.9%, n = 3, P = 0.001).
Intriguingly, the overexpression of EDEM2 or Hrd1 in ninaEG69D−/+ retina significantly restored overall Rh-1 levels detectable through Western blots, when compared to control ninaEG69D−/+ retina (Fig. S5 A and B; n = 4, P = 0.017; n = 4, P < 0.001, respectively). Consistently, EDEM2 or Hrd1 overexpression enhanced the amount of Rh-1 detected in the rhabdomeres of ninaEG69D−/+ flies. We favor the interpretation that under enhanced ERAD activity, misfolded mutant Rh-1 is quickly eliminated to allow more wild-type Rh-1 to fold and avoid degradation, for proper trafficking to rhabdomeres. Collectively, these results show that overexpression of ERAD factors that are sufficient to suppress Rh-1-induced ER stress in larval imaginal discs can delay the course of age-related retinal degeneration in the Drosophila model for ADRP.
Discussion
Here, we report on the mechanism and functional consequence of Rh-1 degradation through ERAD in a ninaE mutant allele that serves as a Drosophila model of ADRP. Specifically, we established a Rh-1 overexpression assay in larval imaginal discs and showed that certain ERAD factors can reduce Rh-1 levels to the extent of abolishing ER stress markers when co-expressed. The imaginal disc assay also revealed an unexpected degree of specificity between specific ERAD regulators and their substrates. Most significantly, the findings in the imaginal disc overexpression assay directly correlated with a given ERAD factor's ability to physically bind its substrates, and to suppress retinal degeneration in a physiologically relevant disease model for ADRP, in which age-related retinal degeneration is caused by the endogenous ninaEG69D allele.
One of the unexpected outcomes of our study is the observed specificity between given ERAD factors and their misfolded protein substrates. Previous studies conducted in yeast led to the proposal of the presence of three ERAD subpathways, defined by the subcellular locations of lesions that cause protein misfolding. In that view, the yeast homolog of EDEM, Htm1p, and membrane proteins, Derlin and Usa1p were considered specific components of the ERAD-L pathway, specializing in ER luminal protein recognition and degradation (13–16). By contrast, our study shows that Drosophila EDEM2, Derlin-1 and Herp are involved in reducing the levels of a mutant membrane protein, Rh-1G69D. The list of ERAD genes that can reduce Rh-1G69D levels were different from those involved in wild-type Rh-1 protein or alpha 1-antitrypsinNHK, which is an established ER luminal substrate. We favor the interpretation that EDEM1 initiates an ERAD-L-like pathway in Drosophila, in part, based on the fact that EDEM1 only reduced alpha 1-antitrypsin but not the Rh-1 proteins. On the other hand, Hrd1 overexpression may initiate an ERAD-M-like pathway, as this condition only affected Rh-1 proteins, but not alpha 1-antitrypsin. The observation that Rh-1G69D degradation properties are neither identical to Rh-1WT nor alpha 1-antitrypsinNHK degradation suggests that animals have evolved additional ERAD subpathways not reported in yeast. Such an idea has been suggested previously (32), but awaits further validation.
While the reductionist approach taken in the eye imaginal disc assay has allowed us to match specific ERAD regulators with wild-type or mutant Rh-1 alleles, we were not able to follow the fates of wild-type and mutant Rh-1 proteins that must exist as a mix in the ninaEG69D−/+ retina. Previous studies have demonstrated that, under such a condition, the mutant Rh-1 proteins interfere with the proper maturation of the wild-type Rh-1, leading to the degradation of both species (4, 7, 8, 22). Based on this together with our imaginal disc overexpression assays, we speculate that EDEM2 or Hrd1 stimulates the degradation of misfolded Rh-1 proteins, thereby allowing more wild-type Rh-1 to undergo maturation. Supporting this idea, we found that stimulating ERAD in the ninaEG69D−/+ retina actually enhanced overall Rh-1 levels and a more efficient Rh-1 trafficking to the rhabdomeres (Fig. S5). Elimination of misfolded Rh-1 that may otherwise cause toxicity, together with enhanced Rh-1 trafficking, most likely contribute to the suppression of retinal degeneration in this Drosophila model for ADRP.
Since the ninaEG69D−/+ flies have a mutation that is molecularly similar to those mutations found in human patients, leading to a similar pattern of late-onset retinal degeneration, the role of ERAD in this disease progression is likely conserved between the two species. We exploited ERAD to delay disease in an animal disease model. As ER stress underlies a wide variety of diseases, manipulation of ERAD may be used to therapeutically intervene in a variety of ER stress-related diseases.
Materials and Methods
Plasmids and Fly Stocks.
The following flies and DNA have been described previously: ninaEG69D/TM6B (8), UAS-xbp1-EGFP, UAS-Rh-1G69D (9), Rh1-Gal4, Rh1-GFP (30), gmr-Gal4 (33), hs-flp; tubulin>FRT>y+, GFP>FRT>Gal4 flies (9), Drob-1 expression plasmid (25) alpha 1-antitrypsinNHK DNA (34). The coding sequences for Hrd1, EDEM1, EDEM2, Hrd3, Herp, Der-1, and Der-2 were obtained through RT-PCR from yw larvae. Myc-tags were added to the N termini of these coding sequences and subcloned into a pUAST (35). pGMR-Rh-1G69D construct was created by subcloning the corresponding cDNA into the pGMR vector (33). HerpG13463 allele was obtained from BMRC KAIST. For in vivo RNAi, UAS-Hrd1-IR (V6870), UAS-Hrd3-IR (V1161), UAS-Herp-IR (V11724, V11725), and UAS-Der-1-IR (V44210, V44211) were obtained from the Vienna Drosophila RNAi Center (http://stockcenter.vdrc.at). To enhance the efficiency of RNAi knockdown, uas-dcr2 was driven for co-expression with these inverted repeat lines. Additional methods are available in SI Text.
Supplementary Material
Acknowledgments.
We thank Kazuhiro Nagata (Kyoto University) and Masayuki Miura (Tokyo University) for plasmids; Pedro Domingos, Alexis Gambis, Iwona Gumper, Young Kwon (Johns Hopkins), and Yihong Ye (NIH) for technical advice; and Milton Adesnik (NYU), David Ron (NYU), and Peter Shapiro (NYU) for comments on the manuscript. This project was supported by grants from the National Institutes of Health (1RO1GM079425) and the Ellison Medical Foundation (to H.D.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905566106/DCSupplemental.
References
- 1.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 2.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2007;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dryja T, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 4.Sung CH, et al. Rhdopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Tousa JE, et al. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:877–882. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 6.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 7.Kurada P, O'Tousa JE. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 8.Colley NJ, Cassill JA, Baker EK, Zuker CS. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc Natl Acad Sci USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JH, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanism of cell death in rhodopsin retinitis pigmentosa: Implications for therapy. Trends Mol Med. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 15.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 16.Quan EM, et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih HP, Hales KG, Pringle JR, Peifer M. Identification of septin-interacting proteins and characterization of the Smt3/SUMO-conjugation system in Drosophila. J Cell Sci. 2002;115:1259–1271. doi: 10.1242/jcs.115.6.1259. [DOI] [PubMed] [Google Scholar]
- 18.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 19.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 20.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 21.Leonard DS, Bowman VD, Ready DF, Pak WL. Degeneration of photoreceptors in rhodopsin mutants of Drosophila. J Neurobiol. 1992;23:605–626. doi: 10.1002/neu.480230602. [DOI] [PubMed] [Google Scholar]
- 22.Rajan RS, Kopito RR. Suppression of wild-type rhodopsin maturation by mutants linked to autosomal dominant retinitis pigmentosa. J Biol Chem. 2005;280:1284–1291. doi: 10.1074/jbc.M406448200. [DOI] [PubMed] [Google Scholar]
- 23.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 24.Yu SY, et al. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 25.Igaki T, et al. Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc Natl Acad Sci USA. 2000;97:662–667. doi: 10.1073/pnas.97.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 27.Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 28.Clerc S, et al. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichaud F, Desplan C. A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development. 2001;128:815–826. doi: 10.1242/dev.128.6.815. [DOI] [PubMed] [Google Scholar]
- 31.Ready DF, Ilanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 32.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:212–219. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 34.Hosokawa N, et al. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.