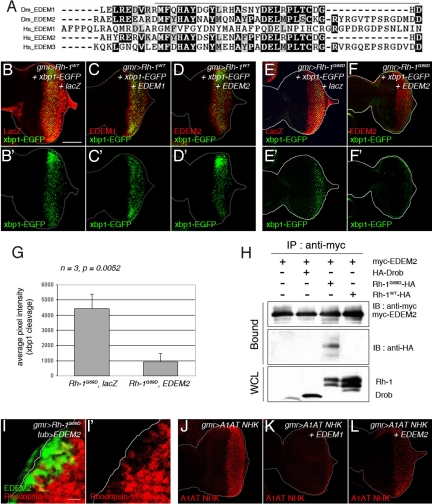
Fig. 3.
Drosophila EDEM2 overexpression reduces mutant, but not wild-type Rh-1 levels. (A) The amino acid sequence alignment between EDEM family proteins of Drosophila and humans, generated with the ClustalW algorithm. Dark shading indicates identity, whereas light shading indicates similarity. Solid lines indicate the catalytic domain of class I mannosidase (Glycosyl hydrolase family 47). The sequences shown, with their corresponding NCBI database accession numbers, are as follows: D. melanogaster (Dm) EDEM1 (NP_726777); D. melanogaster (Dm) EDEM2 (AAF53255); human (Hs) EDEM1 (AAH19088); human (Hs) EDEM2 (NP_060687); human (Hs) EDEM3 (NP_079467). (B–D) The effect of Drosophila EDEM1 and EDEM2 on misexpressed Rh-1WT. Shown are representative discs expressing Rh-1WT, together with lacZ (B), EDEM1 (C), or EDEM2 (D), or expressing Rh-1G69D with lacZ (E) or EDEM2 (F). The degree of ER stress is assessed through xbp1-EGFP activation (B′–F′, green). (G) Quantification of the xbp1-EGFP activation levels as shown in (E and F). Error bars, ± SEM. (H) EDEM2 physically interacts with Rh-1G69D in S2 cells. EDEM2 was immunoprecipitated through its myc-tag, and its interaction partners were detected through their HA-epitopes. (I and I′) A disc expressing EDEM2 (green) in a flip-out mosaic clone shows reduced levels of Rh-1G69D (red). (J–L) Both of EDEM1 and EDEM2 effectively downregulated the level of alpha 1-antitrypsinNHK (A1AT NHK, in red). (Scale bars, 100 μm (B) and 10 μm (I).) Genotypes: gmr-Gal4/UAS-lacZ;UAS-Rh-1WT/UAS-xbp1-EGFP (B and B′), gmr-Gal4/+;UAS-Rh-1WT, UAS-EDEM1/UAS-xbp1-EGFP (C and C′), gmr-Gal4/UAS-EDEM2;UAS-Rh-1WT/UAS-xbp1-EGFP (D and D′), gmr-Gal4/UAS-lacZ;UAS-Rh-1G69D/UAS-xbp1-EGFP (E and E′), gmr-Gal4/UAS-EDEM2;UAS-Rh-1G69D/UAS-xbp1-EGFP (F and F′), hs-flp;UAS-EDEM2/+;tub>GFP>Gal4/gmr-Rh-1G69D (I and I′), gmr-Gal4/UAS-alpha1-antitrypsinNHK;+/+ (J), gmr-Gal4/UAS-alpha1-antitrypsinNHK;UAS-EDEM1/+ (K), and gmr-Gal4/UAS-alpha1-antitrypsinNHK, UAS-EDEM2;+/+ (L).