It is not generally appreciated that descendents of the H1N1 influenza A virus that caused the catastrophic and historic pandemic of 1918-1919 have persisted in humans for more than 90 years and have continued to contribute their genes to new viruses, causing new pandemics, epidemics, and epizootics (see table). The current international pandemic caused by a novel influenza A (H1N1) virus derived from two unrelated swine viruses, one of them a derivative of the 1918 human virus,3 adds to the complexity surrounding this persistent progenitor virus, its descendants, and its several lineages (see diagram).
Table. Mortality Associated with Influenza Pandemics and Selected Seasonal Epidemic Events, 1918-2009*.
| Years | Circulating Virus (Genetic Mechanism) | Excess Deaths from Any Cause no. per 100,000 persons/yr |
|---|---|---|
| 1918-1919 | H1N1 (viral introduction), pandemic | 598.0 |
| 1928-1929 | H1N1 (drift) | 96.7 |
| 1934-1936 | H1N1 (drift) | 52.0 |
| 1947-1948 | H1N1 A’ (intrasubtypic reassortment) | 8.9 |
| 1951-1953 | H1N1 (intrasubtypic reassortment) | 34.1 |
| 1957-1958 | H2N2 (antigenic shift), pandemic | 40.6 |
| 1968-1969 | H3N2 (antigenic shift), pandemic | 16.9 |
| 1972-1973 | H3N2 A Port Chalmers (drift) | 11.8 |
| 1975-1976 | H3N2 (drift) and H1N1 (“swine flu” outbreak) | 12.4 |
| 1977-1978 | H3N2 (drift) and H1N1 (viral return) | 21.0 |
| 1997-1999 | H3N2 A Sydney (intrasubtypic reassortment) and H1N1 (drift) | 49.5 |
| 2003-2004 | H3N2 A Fujian (intrasubtypic reassortment) and H1N1 (drift) | 17.1 |
| 2009 | H3N2 and H1N1 (drift) and swine-origin H1N1 (viral introduction), pandemic | ? |
A useful way to think about influenza A events of the past 91 years is to recognize that we are living in a pandemic era that began around 1918.4 At that time, a presumably new founding virus, containing a novel set of eight influenza genes and probably derived from an unidentified avian like precursor virus, became adapted to mammals; the molecular and virologic events responsible for that adaptation remain unclear. This virus caused an explosive and historic pandemic, during which humans also transmitted the virus to pigs, in which it remains in circulation. Ever since 1918, this tenacious virus has drawn on a bag of evolutionary tricks to survive in one form or another, in both humans and pigs, and to spawn a host of novel progeny viruses with novel gene constellations, through the periodic importation or exportation of viral genes (see Zimmer and Burke, pages 279-285). The 2009 H1N1 pandemic virus represents yet another genetic product in the still-growing family tree of this remarkable 1918 virus.
To understand what has been happening since 1918, it is helpful to think of influenza viruses not as distinct entities but as eight-member “gene teams” that work together and must some times trade away one or more team members to make way for new gene “players” with unique skills. In nature, avian influenza A viruses seem to exist as transient complexes of eight genes that assemble and reassemble promiscuously, if not randomly, in an enormous global avian reservoir. Within this reservoir, avian viruses remain stably adapted to the enteric tracts of hundreds of avian species, single members of which are often simultaneously infected by multiple viruses that engage in prolific gene reassortment. Because of this continual reassortment, a seemingly endless variety of new viruses with potentially new properties are continually being engineered. Indeed, thousands of unique gene constellations making up avian influenza viruses have already been identified; as research continues, the number will undoubtedly grow.
The mechanisms by which avian viruses cross species barriers to infect humans or other mammals, either causing dead-end infections or leading to subsequent human-to-human transmission, are unknown. Moreover, the properties of influenza viruses that have the greatest medical and public health relevance, such as human infectivity, transmissibility, and pathogenicity, appear to be complex and polygenic and are poorly understood. Every influenza A virus has a gene coding for 1 of 16 possible hemagglutinin (HA) surface proteins and another gene coding for 1 of 9 possible neuraminidase (NA) surface proteins. These two proteins (facilitating viral attachment and release, respectively) not only are critical for the infection of susceptible cells of a host but also elicit immune responses that prevent infection or independently reduce viral replication, respectively. Of the 144 total combinatorial possibilities, only three HAs and two NAs, in only 3 combinations (H1N1, H2N2, and H3N2), have ever been found in truly human-adapted viruses — a fact that suggests inherent limitations in host adaptation. In addition to possible constraints related to HA or NA, viruses adapted to humans or other mammals may be constrained by a need for all their genes to be coadapted both to the host and to each other — a requirement that seems to be particularly difficult to fulfill. Chimeric viruses containing fewer than all eight genes of the 1918 virus, for example, are not as pathogenic in animal models as the fully reconstructed 1918 virus.
Once new human influenza viruses appear and cause pandemics, population immunity to their HA and NA proteins increases quickly. The powerful counterforce of population immunity is met by the remarkable ability of influenza virus to evolve by means of mutation (drift) or acquisition through reassortment either of different HA subtypes (shift) or through intrasubtypic reassortment with variant HAs of the same subtype or of other genes of cocirculating viruses.5 Direct descendants of the 1918 virus caused “shift pandemics” in 1957 (H2N2) and 1968 (H3N2); they also caused “pandemic-like events” associated with intrasubtypic reassortment in 1947 (H1N1), 1951 (H1N1), 1997 (H3N2), and 2003 (H3N2). By convention, the term “pandemic” influenza has been reserved for global influenza epidemics caused by viruses with new HA subtypes; it has not been consistently applied to widespread or even global epidemics resulting from other viral genetic changes. But the long-held belief that shifts always cause severe pandemics, whereas drifts lead to more modest increases in seasonal mortality, has been called into question. The effects on mortality of new influenza viruses created by the several genetic mechanisms mentioned above are not easily characterized (see table).1,2 In this regard, it is noteworthy that although the precise viruses that circulated before 1918 and the mechanisms of their generation are unknown, probable influenza pandemics have, over several centuries, shown marked variation in severity, ranging from mild (e.g., the 1761-1762 pandemic) to severe (e.g., the 1833-1837 pandemic, which had a 2% case fatality rate).
Background Reading
Collins SD, Lehmann J. Excess deaths from influenza and pneumonia and from important chronic diseases during epidemic periods, 1918-51. Government Printing Office; Washington, DC: 1953. Dowdle WR. Influenza: epidemic patterns and antigenic variation. In: Selby P, editor. Influenza: virus, vaccine and strategy. Academic Press; New York: 1976. pp. 17–21.Dugan VG, Chen R, Spiro DJ, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4(5):e1000076. doi: 10.1371/journal.ppat.1000076.Foppa IM, Hossain MM. Revised estimates of influenza-associated excess mortality, United States, 1995 through 2005. Emerg Themes Epidemiol. 2008;5:26. doi: 10.1186/1742-7622-5-26.Holmes EC, Ghedin E, Miller N, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3(9):e300. doi: 10.1371/journal.pbio.0030300.Kash JC, Tumpey TM, Proll SC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–81. doi: 10.1038/nature05181.Kilbourne ED, Smith C, Brett I, Pokorny BA, Johansson B, Cox N. The total influenza vaccine failure of 1947 revisited: major intrasubtypic antigenic change can explain failure of vaccine in a post-World War II epidemic. Proc Natl Acad Sci U S A. 2002;99:10748–52. doi: 10.1073/pnas.162366899.Memoli MJ, Jagger BW, Dugan VG, Jackson JP, Taubenberger JK. Recent human influenza A/H3N2 virus evolution driven by novel selection factors in addition to antigenic drift. J Infect Dis. doi: 10.1086/605893. (in press)Nelson MI, Viboud C, Simonsen L, et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008;4(2):e1000012. doi: 10.1371/journal.ppat.1000012.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810.Pappas C, Aguilar PV, Basler CF, et al. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc Natl Acad Sci U S A. 2008;105:3064–9. doi: 10.1073/pnas.0711815105.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–9. doi: 10.1038/nature06945.Reid AH, Taubenberger JK, Fanning TG. Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Nat Rev Microbiol. 2004;2:909–14. doi: 10.1038/nrmicro1027.Simonsen L, Clarke MJ, Stroup DF, Williamson GD, Arden NH, Cox NJ. A method for timely assessment of influenza-associated mortality in the United States. Epidemiology. 1997;8:390–5. doi: 10.1097/00001648-199707000-00007.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979.Taubenberger JK, Reid AH, Janczewski TA, Fanning TG. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos Trans R Soc Lond B Biol Sci. 2001;356:1829–39. doi: 10.1098/rstb.2001.1020.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–93. doi: 10.1038/nature04230.Wang R, Soll L, Dugan V, et al. Examining the hemagglutinin subtype diversity among wild duck-origin influenza A viruses using ethanol-fixed cloacal swabs and a novel RT-PCR method. Virology. 2008;375:182–9. doi: 10.1016/j.virol.2008.01.041.Wolf YI, Viboud C, Holmes EC, Koonin EV, Lipman DJ. Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus. Biol Direct. 2006;1:34. doi: 10.1186/1745-6150-1-34.Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1691–740.
It is remarkable not only that direct “all-eight-gene” descendants of the 1918 virus still circulate in humans as epidemic H1N1 viruses and in swine as epizootic H1N1 viruses, but also that for the past 50 years the original virus and its progeny have continually donated genes to new viruses to cause new pandemics, epidemics, and epizootics. The novel H1N1 virus associated with the ongoing 2009 pandemic is a fourth-generation descendant of the 1918 virus. The complex evolutionary history of this virus features genetic mixing both within human viruses and between avian- and swine-adapted influenza viruses, gene-segment evolution in multiple species, and evolution in response to the selection pressures of herd immunity in various populations at various points in time. The fact that this novel H1N1 influenza A virus has become a pandemic virus expands the previous definition of the term.
The 1918 influenza virus and its progeny, and the human immunity developed in response to them, have for nearly a century evolved in an elaborate dance; the partners have remained linked and in step, even as each strives to take the lead. This complex interplay between rapid viral evolution and virally driven changes in human population immunity has created a “pandemic era” lasting for 91 years and counting. There is little evidence that this era is about to come to an end.
If there is good news, it is that successive pandemics and pandemic-like events generally appear to be decreasing in severity over time. This diminution is surely due in part to advances in medicine and public health, but it may also reflect viral evolutionary “choices” that favor optimal transmissibility with minimal pathogenicity — a virus that kills its hosts or sends them to bed is not optimally transmissible. Although we must be prepared to deal with the possibility of a new and clinically severe influenza pandemic caused by an entirely new virus, we must also understand in greater depth, and continue to explore, the determinants and dynamics of the pandemic era in which we live.
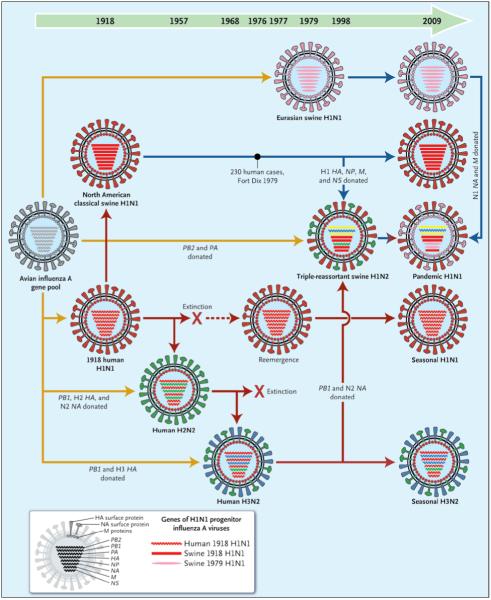
Genetic Relationships among Human and Relevant Swine Influenza Viruses, 1918-2009.
Yellow arrows reflect exportation of one or more genes from the avian influenza A virus gene pool. The dashed red arrow indicates a period without circulation. Solid red arrows indicate the evolutionary paths of human influenza virus lineages; solid blue arrows, of swine influenza virus lineages; and the blue-to-red arrow, of a swine-origin human influenza virus. All influenza A viruses contain eight genes that encode the following proteins (shown from top to bottom within each virus): polymerase PB2, polymerase PB1, polymerase PA, hemagglutinin (HA), nuclear protein (NP), neuraminidase (NA), matrix proteins (M), and nonstructural proteins (NS). The genes of the 1918 human and swine H1N1 and the 1979 H1N1 influenza A viruses were all recently descended from avian influenza A genes, and some have been “donated” to the pandemic human H1N1 strain.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Noble GR. Epidemiological and clinical aspects of influenza. In: Beare AS, editor. Basic and applied influenza research. CRC Press; Boca Raton, FL: 1982. pp. 11–50. [Google Scholar]
- 2.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 3.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009 May 22; doi: 10.1126/science.1176225. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Morens DM. Pandemic influenza — including a risk assessment of H5N1. Rev Sci Tech. 2009;28:187–202. doi: 10.20506/rst.28.1.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–9. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]