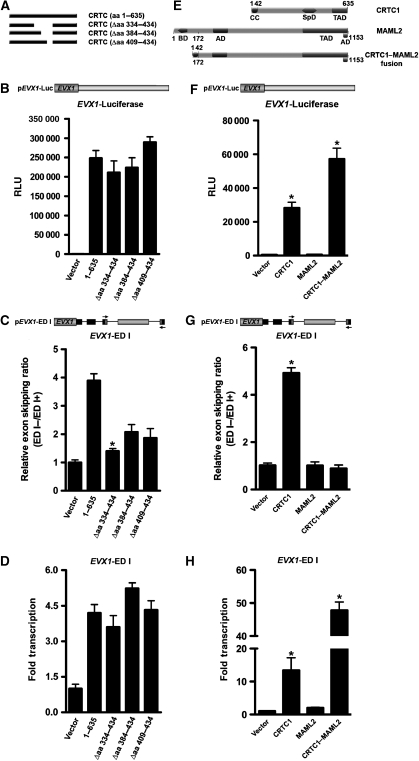
Figure 7.
Transcriptional activation is not required for CRTC-dependent alternative splicing. (A) Schematic illustration of full-length and internal CRTC deletion proteins with corresponding deleted amino acids indicated in parentheses. (B) Transient co-transfection of CRTC deletion mutants with EVX1 luciferase reporter into HEK293T cells were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (C) TaqMan real-time qPCR analysis of CRTC deletion mutants on EVX1 ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (D) TaqMan real-time qPCR analysis of CRTC deletion mutants on EVX1 ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (E) Schematic illustration of key CRTC1 and MAML2 protein domains relative to the CRTC1–MAML2 chimeric oncoprotein. Corresponding amino acids derived from CRTC1 are indicated above, whereas those derived from MAML2 are indicated below. Abbreviations are as follows: AD, acidic domain; BD, basic domain; CC, coiled-coil; SpD, splicing domain; TAD, transactivation domain. (F) Transient co-transfection of CRTC1–MAML2 chimeric oncoprotein with EVX1 luciferase reporter into HEK293T cells were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (G) TaqMan real-time qPCR analysis of CRTC1–MAML2 chimeric oncoprotein on EVX1 ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (H) TaqMan real-time qPCR analysis of CRTC1–MAML2 chimeric oncoprotein on EVX1 ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05).