Abstract
The breast cancer 2, early onset protein (BRCA2) is central to the repair of DNA damage by homologous recombination. BRCA2 recruits the recombinase RAD51 to sites of damage, regulates its assembly into nucleoprotein filaments and thereby promotes homologous recombination. Localization of BRCA2 to nuclear foci requires its association with the partner and localizer of BRCA2 (PALB2), mutations in which are associated with cancer predisposition, as well as subtype N of Fanconi anaemia. We have determined the structure of the PALB2 carboxy-terminal β-propeller domain in complex with a BRCA2 peptide. The structure shows the molecular determinants of this important protein–protein interaction and explains the effects of both cancer-associated truncating mutants in PALB2 and missense mutations in the amino-terminal region of BRCA2.
Keywords: PALB2, BRCA2, DNA repair, homologous recombination, cancer
Introduction
Repair of DNA double-strand breaks and interstrand crosslinks by homologous recombination in human cells involves the co-ordinated assembly of several multi-protein complexes. Central to these is the scaffold protein BRCA2 (breast cancer 2, early onset), which regulates the assembly of RAD51 recombinase into nucleoprotein filaments (Sharan et al, 1997; Yuan et al, 1999). Heterozygous germline defects in BRCA2 predispose a person to breast and ovarian cancers, whereas homozygous defects cause Fanconi anaemia (Howlett et al, 2002; Gudmundsdottir & Ashworth, 2006). Cells with BRCA2 defects show genetic instability, consistent with abrogation of homologous recombination, and are typified by chromosomal rearrangements and aneuploidy (Patel et al, 1998; Tutt et al, 1999; Daniels et al, 2004).
BRCA2 function depends to a large extent on partner and localizer of BRCA2 (PALB2; also known as FANC-N, for Fanconi anaemia subtype N), for recruitment to nuclear foci and for much of its recombinational activity (Xia et al, 2006). As with BRCA2, heterozygous germline mutations in PALB2 are associated with cancer predisposition, and homozygous germline mutations result in a form of Fanconi anaemia (Rahman et al, 2007; Reid et al, 2007; Xia et al, 2007). Recently, PALB2 mutations have also been associated with a susceptibility to pancreatic cancer (Jones et al, 2009). PALB2 is a 130-kDa protein with no clear functional domains other than a predicted amino-terminal coiled-coil structure and a carboxy-terminal WD40-repeat motif. Deletion mapping has localized the PALB2-interacting region of BRCA2 to residues 10–40 at the N-terminus of the protein (Xia et al, 2006).
We have determined the structure of the C-terminal region of PALB2, showing a seven-bladed WD40-type β-propeller, which provides the binding site for the N-terminus of BRCA2. The structure of the core PALB2–BRCA2 complex shows a new interaction with a pocket on the face of the structure opposite to that seen in most WD40–peptide interactions. Together, these structures explain the effects of cancer-associated truncating mutations in PALB2 and of missense mutations in the N-terminal region of BRCA2. The interaction revealed by our structural studies is found to be both necessary and sufficient for the association of PALB2 and BRCA2 in cells.
Results And Discussion
Structure of the PALB2 carboxy-terminal domain
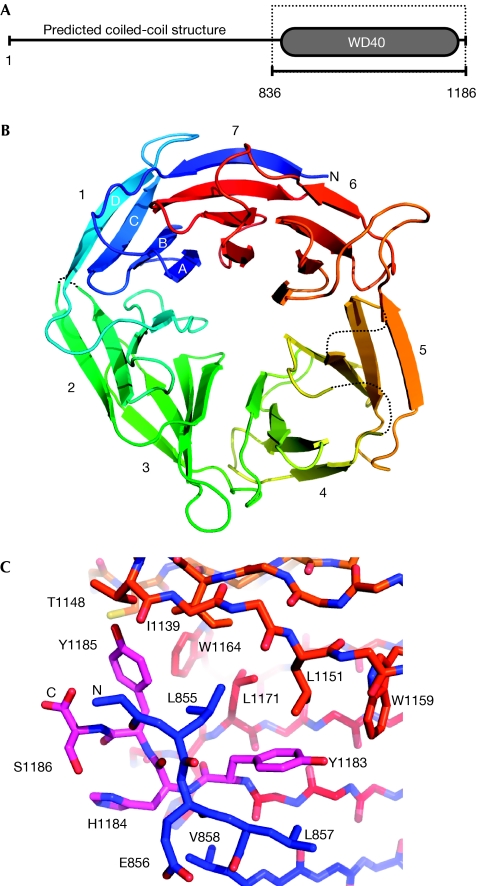
Truncating mutations in PALB2 are associated with cancer predisposition (Rahman et al, 2007; Reid et al, 2007; Tischkowitz et al, 2007; Xia et al, 2007), implicating the C-terminus in mediating interaction with BRCA2. We defined a C-terminal region in PALB2 that had the characteristics of a seven-bladed β-propeller—a domain commonly involved in protein–protein interactions—and a construct encoding this region (PALB2-C; Fig 1A) was expressed in Sf9 cells, purified and crystallized, and the structure determined by X-ray crystallography (see Methods, supplementary information online and Table 1).
Figure 1.
Structure of the PALB2 carboxy-terminal domain. (A) Bioinformatics analysis identifies a WD40-repeat domain forming the C-terminal part of PALB2 whereas the N-terminal and central regions are predicted to contain a coiled-coil structure. The boxed region indicates the amino-acid boundaries of the PALB2-C expression construct used in this study. (B) PALB2-C is a seven-bladed β-propeller (rainbow coloured blue → red, N → C terminus). Blade 7 is formed by strands A, B and C from the C-terminus and strand D from the N-terminus, which seal the toriodal structure in a ‘molecular Velcro' interaction. (C) Details of strand C of blade 7 and its environment. In families with Fanconi anaemia that have Y1183X mutations, the absence of C-terminal residues (magenta) prevents the closure of the WD40 ring and destabilizes the PALB2 protein, which is undetectable in these patients. PALB2, partner and localizer of BRCA2.
Table 1.
Data collection, phasing and refinement statistics
| PALB2+KAu(CN)2 | PALB2 native (2W18) | PALB2+peptide (3EU7) | |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 81.52, 63.15, 77.48 | 80.78, 64.65, 77.25 | 82.82, 62.03, 77.98 |
| α, β, γ (°) | 90, 109.69, 90 | 90, 109.65, 90 | 90, 108.23, 90 |
| Resolution (Å) | 40.0–2.1 (2.2–2.1)* | 49.3–1.9 (2.0–1.9) | 37.5–2.1 (2.32–2.1) |
| Rmerge | 0.06 (0.41) | 0.06 (0.58) | 0.07 (0.60) |
| Mn I/σI | 17.6 (4.2) | 11.2 (2.7) | 16.5 (2.9) |
| Completeness (%) | 96.3 (80.6) | 94.1 (75.4) | 99.9 (99.0) |
| Redundancy | 7.6 (7.2) | 4.0 (3.8) | 5.8 (5.9) |
| Refinement | |||
| Resolution (Å) | 49.3–1.9 | 37.5–2.2 | |
| Number of reflections | 27,818 (3238) | 19,026 (2800) | |
| Rwork/Rfree | 0.21/0.25 | 0.20/0.25 | |
| Number of atoms | |||
| Protein | 2,326 | 2,367 | |
| Ligand/ion | 30 (glycerol) | 12 (glycerol) 108 (peptide) | |
| Water | 187 | 98 | |
| B-factors | |||
| Protein | 35.9 | 47.6 | |
| Ligand/ion | 43.1 (glycerol) | 54.9 (glycerol) 55.4 (peptide) | |
| Water | 41.4 | 47.8 | |
| r.m.s.d. values | |||
| Bond lengths (Å) | 0.012 | 0.014 | |
| Bond angles (°) | 1.47 | 1.63 | |
| *Highest resolution shell is shown in parentheses. | |||
PALB2-C has the linear topology seen in other WD40-repeat domains and the toroidal structure ‘sealed' in the seventh blade by interaction of the C-terminal strand with the incomplete N-terminal blade (Fig 1B). The β-propeller structure of PALB2-C provides an explanation for the cancer-associated mutation (3459C → G; Y1183X) that removes the last four residues (Reid et al, 2007). The absence of these residues completely disrupts the ‘molecular Velcro' hydrogen bonding in the seventh blade (Fig 1C), leaving an incompletely folded protein that is likely to be degraded rapidly. Indeed, PALB2 protein is not detectable in lymphoblastoid cells derived from patients with this mutation (Reid et al, 2007).
Interaction of PALB2 with BRCA2
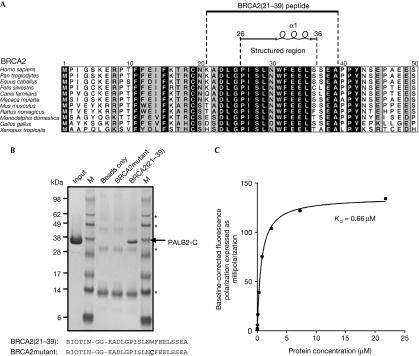
Previous studies localized the PALB2-interacting region of BRCA2 to amino acids 10–40, in which cancer-associated missense mutations have also been observed (Fig 2A). In particular G25R, W31C and W31R result in loss of BRCA2 binding by PALB2 (Xia et al, 2006). We found that a peptide containing residues 21–39 of BRCA2 could readily precipitate PALB2-C in vitro, whereas a peptide containing the W31C mutation could not (Fig 2B), confirming that the C-terminus of PALB2 contains the BRCA2-binding site. Quantitative analysis of this interaction (Fig 2C) gave a dissociation constant (Kd) of ∼0.66 μM. On the basis of this observation, co-crystals of PALB2-C and the wild-type peptide BRCA2(21–39) were obtained, and diffraction data collected and refined against the structure of the ligand-free protein (see Methods). Clear difference electron density for the BRCA2(21–39) peptide was evident and residues 26–36 could be readily fitted and refined (see supplementary Fig S1 online).
Figure 2.
A minimal PALB2-binding motif in the BRCA2 amino terminus. (A) The previously localized PALB2-binding site in residues 10–40 of BRCA2 (Xia et al, 2006) was mapped to residues 21–39, encapsulating a minimal ten-residue motif, which is structured in the PALB2-C–BRCA2(21–39) complex. (B) Biotinylated wild-type BRCA2(21–39) peptide precipitates the PALB2-C construct, confirming it as the BRCA2-binding site. A peptide containing a breast-cancer-associated W31C mutation failed to co-precipitate PALB2-C. Bands marked with an asterisk arise from the NeutrAvidin resin. (C) Fluorescence polarization binding isotherm for interaction of PALB2-C with fluorescein-labelled BRCA2(21–39) peptide (see Methods). Kd as determined by nonlinear fitting of the data with a one-site-specific binding model (GraphPad Prism 5.0). BRCA2, breast cancer 2, early onset; PALB2, partner and localizer of BRCA2.
In many observations of peptide binding to β-propellers, the interaction centres on the mouth of the axial channel opening onto the face containing the loops connecting consecutive ‘blades' (Lodowski et al, 2003; Han et al, 2006; Jennings et al, 2006; Hao et al, 2007; Larsen et al, 2007). However, this is not universal, and interactions with the outer cylindrical surface are observed in the binding of β-adaptin to the N-terminal WD40 domain of clathrin (ter Haar et al, 2000), or in the binding of histone H4 to RbAp46 (Murzina et al, 2008).
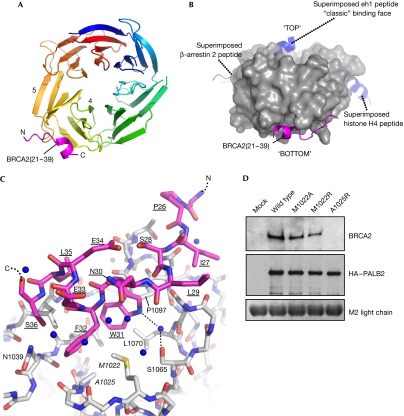
BRCA2(21–39) binds in a pocket formed by the tips of the fourth and fifth blades, on the face opposite to the common axial site (Fig 3A–C). The core of the interaction is provided by BRCA2 Trp31, Phe32 and Leu35, which project from a short helix into a hydrophobic pocket lined by PALB2 residues Val1019, Met1022, Ala1025, Ile1037, Leu1046, Lys1047, Leu1070, Pro1097 and Lys1098. Hydrophobic interactions N-terminal to the helix involve BRCA2 Ile27 and Leu29 and PALB2 Met1067, Gly1068 and Leu1069. The core interface is supported by polar interactions, including a water bridge from the indole nitrogen of BRCA2 Trp31 to Ser1065 in the wall of the PALB2 pocket.
Figure 3.
Structure of PALB2-C–BRCA2(21–39) complex. (A) BRCA2(21–39) (magenta) binds across a hydrophobic pocket at the crossover between blades 4 and 5 of the PALB2-C β-propeller, forming a short α-helix. The r.m.s.d. between the apo-bound and peptide-bound PALB2-C structures is 0.329 Å. (B) Molecular surface of PALB2-C (grey) highlighting the location of BRCA2 binding (magenta) compared with other β-propeller–peptide complexes. The superimposed eh1 motif from a complex with the WD40 domain of TLE1 (Jennings et al, 2006) marks the ‘classic' peptide motif-binding site found in most other WD40–peptide complexes. The positions of β-arrestin 2 and histone H4 peptides (blue), from complexes with the WD40 domains of clathrin and RBBP7, respectively (ter Haar et al, 2000; Murzina et al, 2008), are known examples of non-canonical sites, but are distinct from that observed for the PALB2-C–BRCA2(21–39) complex. (C) Details of the PALB2-C–BRCA2(21–39) interface. The core of the interaction is provided by Trp 31, Phe 32 and Leu 35 inserting into a hydrophobic pocket on PALB2-C (see text). The complex buries ∼1,100 Å2 of the molecular surface, typical of a reversible regulatory interaction. (D) Immunoprecipitation of FLAG–HA-tagged wild-type PALB2 and PALB2 with missense mutations in residues contributing to the BRCA2-binding site (see Methods). Endogenous BRCA2 is precipitated by wild-type PALB2, and to a lesser degree by the Met 1022 mutants. However, BRCA2 binding is abolished by mutation of Ala 1025, which lies at the bottom of the PALB2-hydrophobic pocket. The light chain of the M2 FLAG antibody provides a loading control. BRCA2, breast cancer 2, early onset; HA, haemagglutinin; PALB2, partner and localizer of BRCA2; TLE1, transducin-like enhancer of split 1.
The involvement of BRCA2 Trp31 explains why cancer-associated W31C and W31R mutations disrupt the functional interaction with PALB2. For PALB2 itself, only truncation mutants are described in clinical samples, and these lead to complete loss of the protein. To validate the observed BRCA2-binding site, we generated point mutants in full-length PALB2, designed to disrupt the interaction with BRCA2(21–39), transfected these into human embryonic kidney 293T (HEK-293T) cells, and determined their ability to co-immunoprecipitate endogenous BRCA2 (Fig 3D). Mutation of PALB2 Met1022, on the side of the hydrophobic pocket, only slightly decreased the BRCA2 co-immunoprecipitation. However, mutation of PALB2 Ala1025 at the bottom of the pocket, and in contact with BRCA2 Phe32 in the complex, completely abolished BRCA2 binding, confirming the functional role of the pocket. All the PALB2 mutants were expressed at a level comparable with wild type, and the full-length protein could also be immunoprecipitated at comparable levels, showing that the mutations did not disrupt PALB2 stability. As single missense mutations in either protein can disrupt their interaction, this demonstrates that this interface is both necessary and sufficient for the interaction of BRCA2 and PALB2 in cells.
PALB2 is upstream from BRCA2 in the informational ‘flow' from recognition of a DNA double-strand break to assembly of the RAD51 nucleoprotein filament. The interaction we have characterized mediates the recruitment of BRCA2, but unless PALB2 proves to be able to recognize DNA damage directly, it must also interact with other proteins upstream in the DNA-damage information flow. One recently identified possibility is BRCA1, which seems to interact with N-terminal regions of PALB2 (Zhang et al, 2009). The surface conservation in PALB2-C (supplementary Figs S2 and S3 online) suggests that, apart from the BRCA2 interaction involving the new ‘bottom-face' site, the β-propeller might simultaneously provide interactions through the conventional ‘top-face' site at the mouth of the axial channel. The structure-guided PALB2 missense mutants we have developed here, which specifically abrogate BRCA2 interaction without destabilizing the protein, will be invaluable for identifying further PALB2-interacting proteins that mediate the recruitment of the BRCA2 HR repair complex to sites of DNA damage.
Role of the BRCA2 amino-terminus
BRCA2 has a central role in regulating DNA double-strand break and inter-strand crosslink repair through homologous recombination. A significant component of that role is provided by the regulatory and scaffolding interaction of BRCA2 with the RAD51 recombinase (Thorslund & West, 2007), which is mediated by the central BRC repeat and C-terminal regions of BRCA2 (Saeki et al, 2006; Davies & Pellegrini, 2007; Esashi et al, 2007). Most of the defined pathogenic familial mutations in BRCA2 cause frameshift or nonsense mutations in the BRCA2 open-reading frame, resulting in the expression of truncated or unstable BRCA2 proteins, which are impaired in RAD51 interactions (Wooster et al, 1995; Tavtigian et al, 1996). The critical involvement of the C-terminal RAD51-binding domain, in particular, is highlighted by the ability of reverted mutant BRCA2 proteins that contain substantial internal deletions to restore homologous recombination in BRCA2-defective cancer cell lines (Edwards et al, 2008; Sakai et al, 2008).
The role of the extreme N-terminal region of BRCA2 has been more controversial. Previous studies suggested an involvement in transcriptional activation (Milner et al, 1997; Fuks et al, 1998), a conjecture reinforced by the discovery of EMSY, a protein able to interact with chromatin-regulatory proteins, as a binding partner for the BRCA2 N-terminus (Hughes-Davies et al, 2003). However, the mechanistic implications of possible BRCA2 involvement in transcriptional regulation have not been defined, and the biological significance of this suggestion remains uncertain. The consequences of BRCA2 defects are, however, consistently explained by a primary role in the arrangement and regulation of DNA repair by homologous recombination (Gudmundsdottir & Ashworth, 2006). The discovery of PALB2 as a crucial factor in localizing BRCA2 to sites of DNA damage (Xia et al, 2006) provides a strong argument that the main role of the N-terminal region of BRCA2 is to provide a binding site for PALB2. In support of this argument, we found that the BRCA2 residues we identified as being essential for PALB2 binding are totally conserved in organisms (vertebrates) that have a PALB2 orthologue, but not in those that do not. Although the interaction between PALB2 and BRCA2 has now been characterized at the atomic level, the crucial questions about how PALB2 directs BRCA2 to sites of DNA damage, and the identities of other PALB2-interacting proteins that might be involved, remain to be answered.
Speculation
The PALB2-binding site on BRCA2 directly overlaps that of EMSY, a protein amplified and overexpressed in many breast and ovarian cancers (Hughes-Davies et al, 2003). Tumours with EMSY amplifications and a cell line overexpressing EMSY show chromosomal instability and mitomycin-C sensitivity characteristic of cells defective in BRCA2 or PALB2 (Turner et al, 2004; Raouf et al, 2005; Xia et al, 2006). However, a role for EMSY in BRCA2-mediated homologous recombination or a mechanism by which EMSY overexpression affects the BRCA2 function has not been described. As the PALB2- and EMSY-binding sites on BRCA2 overlap, and preliminary data (supplementary Fig S3 online) indicate mutually exclusive binding, we speculate that high EMSY protein levels due to amplification and/or overexpression in tumours might outcompete PALB2, disrupting the functionally important PALB2–BRCA2 interaction in homologous recombination and engendering the observed genetic instability.
Methods
Peptides. All peptides were chemically synthesized and purified at Peptide Protein Research Ltd, Fareham, UK.
Cloning, expression and purification. PALB2-C (residues 835–1186) was expressed in Sf9 insect cells and purified using immobilized metal-affinity and size-exclusion chromatography. Complete details are provided in the supplementary information online.
Crystallization, phasing and data refinement. PALB2-C was crystallized by vapour diffusion in hanging drops, at a protein concentration of 10 mg/ml and a temperature of 20 °C, using a precipitant containing 100 mM 2-[N-morpholino]ethanesulphonic acid pH 6.0, 50 mM KH2PO4 and PEG 8000 at concentrations between 12 and 20% (wt/vol). Crystals were cryoprotected for data collection by stepwise transfer, to a final concentration of 30% glycerol (vol/vol). A heavy-atom derivative was obtained by soaking native crystals overnight in a stabilizing buffer containing 10 mM KAu(CN)2, before freezing. Co-crystals with the BRCA2(21–39) peptide were grown under similar conditions, with an ∼1:4 ratio of protein:peptide.
All diffraction data were collected on station ID14.1 at the European Synchrotron Radiation Facility, Grenoble, France, and the native PALB2-C structure was phased using single-wavelength anomalous dispersion from the KAu(CN)2 derivative crystals. The native structure was built from Fourier maps and refined to 1.9 Å. This model was then used to phase the PALB2-C–BRCA2(21–39) co-crystals, and the structure of the complex was refined to 2.1 Å. Complete experimental details are provided in the supplementary information online, and statistics for the crystallography are provided in Table 1.
Peptide binding and competition assays. Experiments were carried out with biotinylated peptides corresponding to either residues 21–39 of human BRCA2 (biotin–GG–KALDGPISLNWFEELSSEA) or the same sequence containing a single point mutant W31C (biotin–GG–KALDGPISLNCFEELSSEA). The affinity of BRCA2(21–39) for PALB2-C was determined by fluorescence polarization measurements using an N-terminally fluorescein-labelled wild-type BRCA2(21–39) peptide. Complete details are provided in the supplementary information online.
Mammalian expression and M2-agarose immunoprecipitation. Single point mutants of full-length PALB2, in the mammalian expression vector pOZ-FH-C, were generated using a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) following the manufacturer's recommended protocol. HEK-293T cells were obtained from the American Type Culture Collection (ATCC; LGC Standards, Teddington, UK) and maintained in DMEM containing 10% (vol/vol) fetal calf serum. Cells were transfected in six-well plates using FUGENE6 (Roche, Burgess Hill, UK), according to the manufacturer's instructions. Whole-cell extracts were prepared in 1 ml of NETN420 buffer (20 mM Tris–HCl (pH 7.5), 420 mM NaCl, 1 mM EDTA, 0.5% (vol/vol) IGEPAL CA-630) as described in Xia et al (2006), 48 h after transfection. FLAG-tagged PALB2 was precipitated from these extracts with anti-FLAG M2-agarose beads (Sigma-Aldrich, Gillingham, UK), for a period of 20 h at 4°C. Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to D. Livingston (Dana-Farber Cancer Institute) for the pOZC-PALB2 mammalian expression vector, to K. Wood and A. Ali (Institute of Cancer Research) for assistance with baculovirus expression and protein purification, and to B. Xia (The Cancer Institute of New Jersey), R. Brough (Breakthrough) and S.M. Roe (Institute of Cancer Research) for advice and assistance. This work was supported by Cancer Research UK (Programme Grant C302/A8265 and Infrastructure Support Grant C302/A7803 to LHP) and Breakthrough Breast Cancer. We acknowledge the National Health Service funding to the National Institute for Health Research Biomedical Research Centre. Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (http://www.pdb.org) under the accession codes 2W18 and 3EU7.
Footnotes
The authors declare that they have no conflict of interest.
References
- Daniels MJ, Wang Y, Lee M, Venkitaraman AR (2004) Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science 306: 876–879 [DOI] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L (2007) Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol 14: 475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC (2007) Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol 14: 468–474 [DOI] [PubMed] [Google Scholar]
- Fuks F, Milner J, Kouzarides T (1998) BRCA2 associates with acetyltransferase activity when bound to P/CAF. Oncogene 17: 2531–2534 [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A (2006) The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25: 5864–5874 [DOI] [PubMed] [Google Scholar]
- Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J (2006) Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell 22: 137–144 [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP (2007) Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Howlett NG et al. (2002) Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609 [DOI] [PubMed] [Google Scholar]
- Hughes-Davies L et al. (2003) EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 115: 523–535 [DOI] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D (2006) Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell 22: 645–655 [DOI] [PubMed] [Google Scholar]
- Jones S et al. (2009) Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NA, Al-Bassam J, Wei RR, Harrison SC (2007) Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci USA 104: 1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ (2003) Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science 300: 1256–1262 [DOI] [PubMed] [Google Scholar]
- Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T (1997) Transcriptional activation functions in BRCA2. Nature 386: 772–773 [DOI] [PubMed] [Google Scholar]
- Murzina NV et al. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16: 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR (1998) Involvement of Brca2 in DNA repair. Mol Cell 1: 347–357 [DOI] [PubMed] [Google Scholar]
- Rahman N et al. (2007) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 39: 165–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Brown L, Vrcelj N, To K, Kwok W, Huntsman D, Eaves CJ (2005) Genomic instability of human mammary epithelial cells overexpressing a truncated form of EMSY. J Natl Cancer Inst 97: 1302–1306 [DOI] [PubMed] [Google Scholar]
- Reid S et al. (2007) Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39: 162–164 [DOI] [PubMed] [Google Scholar]
- Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, Han M, Zdzienicka MZ, Stark JM, Jasin M (2006) Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci USA 103: 8768–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W et al. (2008) Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV et al. (1996) The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 12: 333–337 [DOI] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T (2000) Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA 97: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, West SC (2007) BRCA2: a universal recombinase regulator. Oncogene 26: 7720–7730 [DOI] [PubMed] [Google Scholar]
- Tischkowitz M et al. (2007) Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA 104: 6788–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A (2004) Hallmarks of ′BRCAness′ in sporadic cancers. Nat Rev Cancer 4: 814–819 [DOI] [PubMed] [Google Scholar]
- Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A (1999) Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol 9: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792 [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Xia B et al. (2007) Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39: 159–161 [DOI] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY (1999) BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 59: 3547–3551 [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X (2009) PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information