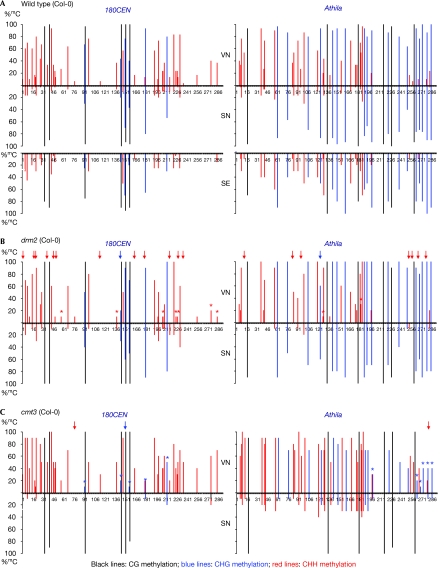
Figure 2.
Bisulphite sequencing analysis of cytosine methylation in wild-type (Col-0), drm2 and cmt3 lines. The graphs show the percentage of methylation (%mC) at individual cytosines in cloned 180CEN (left) and Athila (right) sequences from (A) sorted wild-type vegetative (top) and sperm (middle) nuclei, and seedings (bottom); (B) sorted drm2; and (C) cmt3 vegetative (top) and sperm (bottom) nuclei. Black lines, CG methylation; blue lines, CHG methylation; and red lines, CHH methylation. We define ‘hypermethylated non-CG sites in VN in pollen' as the CHH or CHG sites that are methylated in more cloned DNA sequences from VN than from SN. Note that all CHH sites in both the 180CEN (39 sites) and Athila (30 sites) sequences are hypermethylated in the wild-type VN. Red and blue arrows above the methylation lines indicate the CHH and CHG sites, respectively, at which hypermethylation is lost in mutant VN compared with wild-type VN. Red and blue asterisks mark the CHH and CHG sites, respectively, showing more than a 50% reduction in %mC in mutant VN compared with wild-type VN. Cytosine positions are indicated by numbers. The results are from 30 and 20 cloned sequences for wild-type pollen nuclei and seedlings, respectively, and 10 cloned sequences for drm2 and cmt3 pollen nuclei. Original data are shown in supplementary Fig S4 online. Cmt3, chromomethylase 3; drm2, domains rearranged methyltransferase 2; SE, seedlings; SN, sperm nuclei; VN, vegetative nuclei.