Figure 2.
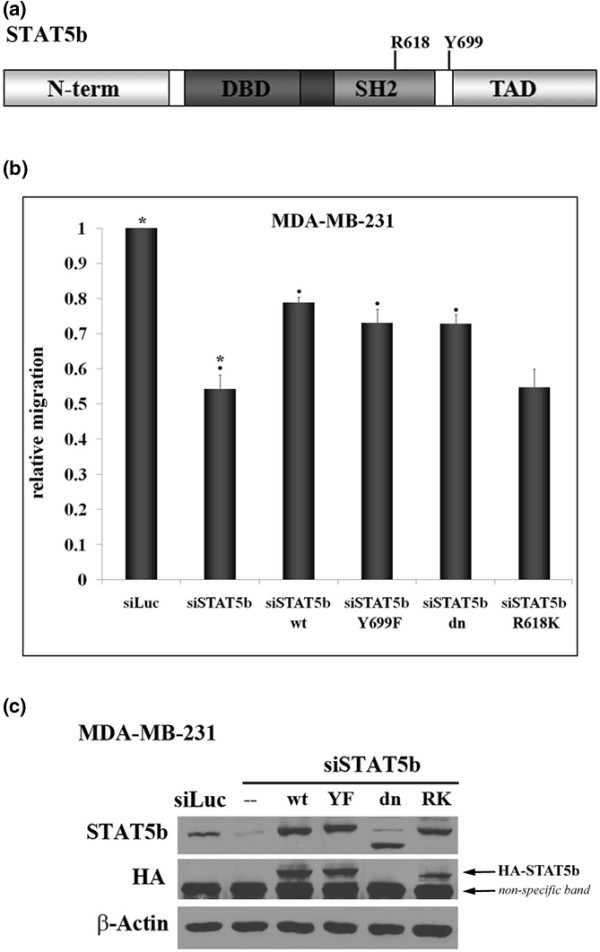
Expression of wild-type, Y699F-, or dominant-negative STAT5b rescues migration, but expression of R618K-STAT5b does not. (a) Domain structure of the STAT5b protein depicting the amino terminus (N-term), DNA-binding domain (DBD), Src homology 2 domain (SH2), transactivation domain (TAD), and the location of the conserved tyrosine residue, Y699, and arginine residue, R618. (b) MDA-MB-231 breast cancer cells were transfected with control siRNA to luciferase (siLuc) or siRNA specific to STAT5b (siSTAT5b siGENOME SMARTpool oligonucleotide #3) alone or in the presence of HA-tagged wild-type (wt-STAT5b), Y699F (YF-STAT5b), dominant-negative (dn-STAT5b), or R618K (RK-STAT5b) STAT5b constructs engineered to be immune to siRNA knockdown. Seventy-two hours after transfection, trans-well assays were performed for 6 hours, as described in Figure 1. Results are graphed as relative migration, compared with siLuc control. One-way ANOVA with Tukey's post-test was used to determine statistical significance (p < 0.05) between the following: siLuc and siSTAT5b (*), siSTAT5b and siSTAT5b + wt-STAT5b (black circles), siSTAT5b and siSTAT5b + YF-STAT5b (black circles), siSTAT5b and siSTAT5b + dn-STAT5b (black circles); n ≥ 4. Comparison of siSTAT5b and siSTAT5b + RK-STAT5b was not statistically significant. Whole-cell lysates from transfected cells were immunoblotted with antibodies specific for STAT5b, HA, or β-actin as a loading control.
