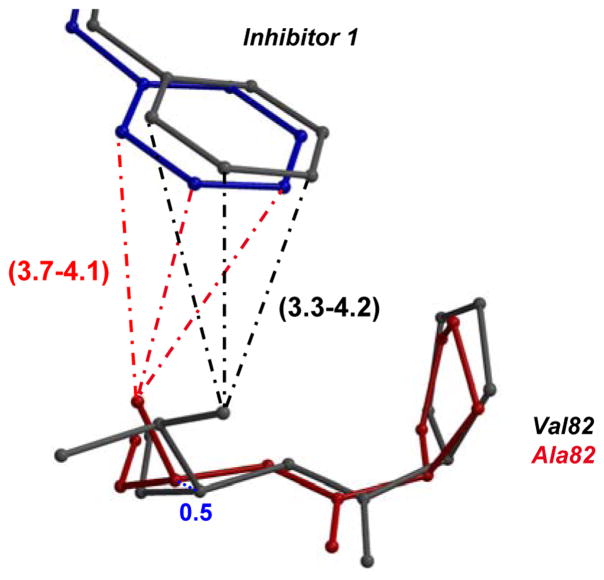
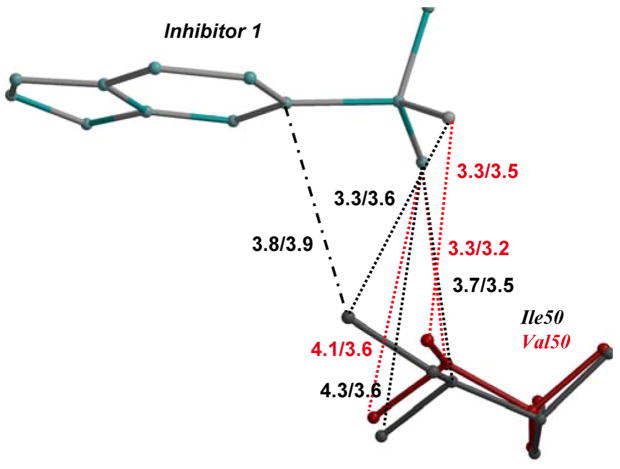
Figure 4. Structural changes in mutants compared to wild type PR.
The major and minor conformations of side chains in PR are shown in dark and light grey ball-and-stick, respectively. The alternate conformations in mutant complexes are shown in red and pink, and the alternate conformations of inhibitor are in blue and cyan. The polar interactions are indicated by the dashed lines, van der Waals interactions by the dotted lines, and C-H…π interactions in dot-and-dash line. The interatomic distances are shown in Å for both subunits.
4A. Interactions of P1′ of inhibitor 1 with residue 82′in PRV82A and PR. The minor conformation of inhibitor 1 and alternate conformation of Val82 in PR are omitted for clarity. The shift in position of Cα 82 between PR and mutant is shown in blue dotted lines. 4B. Van der Waals interactions of P1′ in inhibitor 1 with residue 82′ in PRV82A and PR. View and representation are similar to 4A.
4C. PRI84V and PR interactions with inhibitor 1.
4D. PRI50V and PR interactions with inhibitor 1. Only a single conformation is shown for clarity.
4E. PRD30N and PR interactions with inhibitor 1.
4F. PRD30N and PR interactions with darunavir.