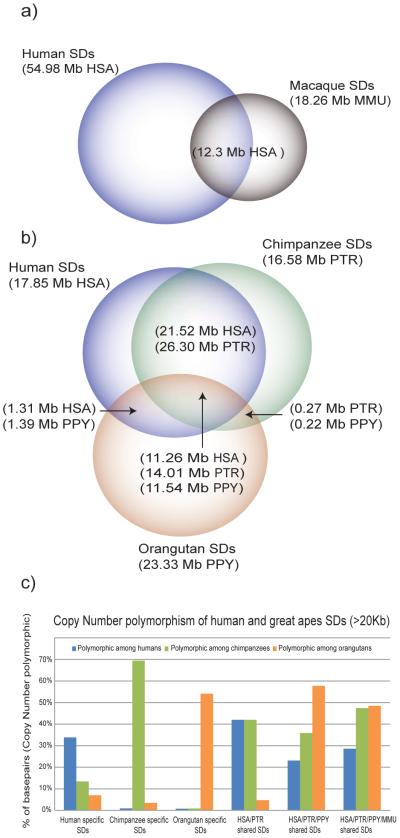
Fig. 2. Shared vs. lineage-specific duplications and great-ape polymorphism.
Segmental duplications (>20 kbp) were classified as lineage-specific or shared based on a four-way comparison of human, chimpanzee, orangutan and macaque genomes. a) Human SDs were compared to Old World monkey SDs (based on a separate analysis of the macaque assembly9). b). Since nonhuman great-ape duplications were detected based on alignment of WGS sequence against the human genome, we corrected for copy number based on the depth of coverage of WGS sequence (in brackets with the name of the species for which the correction was performed, see Table 1). b) Copy-number polymorphic regions were estimated from the results of arrayCGH hybridizations between 8 samples each of human, chimpanzee and orangutan (using the same reference as the computational prediction). The proportion of duplicated bases that showed evidence of copy-number polymorphism (i.e. gain or loss for ≥ two individuals within each species) is shown for each class of SD.