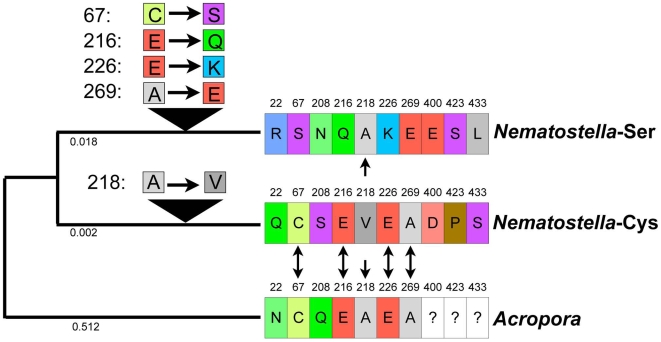
Figure 3. Inferred evolution of polymorphic positions.
A partial coding sequence of the coral Acropora millepora was used to root a phylogenetic tree relating the two Nematostella alleles. A neighbor-joining tree was constructed using the computer program Phylip (version 3.6). Distances between sequences were computed using the first 330 amino acids of the alignment (shown in Figure S3) and the JTT distance matrix. Numbers below branches indicate phylogenetic distance (in units of expected number of substitutions per residue). Positions that are polymorphic in Nematostella are shown to the right. The Acropora sequence was identical to one of the two Nematostella variants at five positions (double-headed arrows). For each of these five positions, substitutions were mapped to either the branch leading to the Ser allele or the branch leading to the Cys allele, assuming that the condition found in Acropora is the ancestral state.