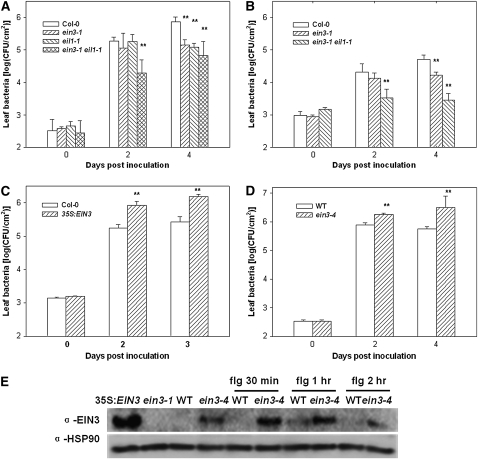
Figure 2.
EIN3 and EIL1 Negatively Regulate Resistance to P. syringae Bacteria.
(A) ein3-1 eil1-1 exhibits enhanced resistance to DC3000. Plants of the indicated genotypes were inoculated with DC3000 bacteria, and bacterial populations in the leaf were determined at the indicated times.
(B) ein3-1 eil1-1 shows enhanced resistance to DC3000 (avrRpt2). Plants of the indicated genotypes were inoculated with DC3000 (avrRpt2) bacteria, and bacterial populations in the leaf were determined at the indicated times.
(C) EIN3 overexpression enhances resistance to DC3000. Plants of the indicated genotypes were inoculated with DC3000 bacteria, and bacterial populations in the leaf were determined at the indicated times.
(D) ein3-4 enhances susceptibility to DC3000. Wild-type RAP2.6-LUC transgenic plants were used as a control.
(E) ein3-4 enhances EIN3 protein stability. Four-day-old etiolated seedlings were treated with 1 μM flg22 for the indicated times and examined by immunoblot using anti-EIN3 antibodies or anti-HSP90 antibodies (for loading control). EIN3 protein exists at a low level in wild-type RAP2.6-LUC transgenic seedlings and was not detected in this blot. The 35S:EIN3 and ein3-1 seedlings were used as positive and negative controls, respectively.
Each data point consisted of at least three samples. Error bars indicate sd. * and ** indicate significant difference at 0.05 and 0.01, respectively, between mutants and Col-0 or wild-type RAP2.6-LUC plants at the same time point (Student's t test). Results shown are a representative of three ([A] to [C]) or five (D) independent experiments.