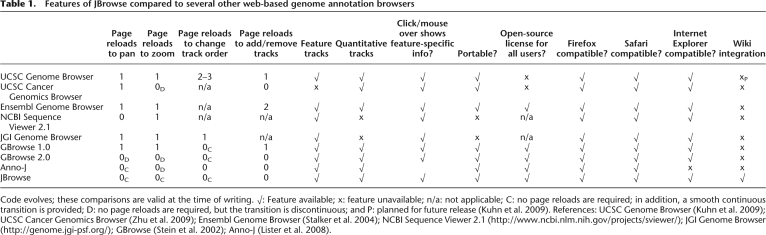
Table 1.
Features of JBrowse compared to several other web-based genome annotation browsers
Code evolves; these comparisons are valid at the time of writing. √: Feature available; x: feature unavailable; n/a: not applicable; C: no page reloads are required; in addition, a smooth continuous transition is provided; D: no page reloads are required, but the transition is discontinuous; and P: planned for future release (Kuhn et al. 2009). References: UCSC Genome Browser (Kuhn et al. 2009); UCSC Cancer Genomics Browser (Zhu et al. 2009); Ensembl Genome Browser (Stalker et al. 2004); NCBI Sequence Viewer 2.1 (http://www.ncbi.nlm.nih.gov/projects/sviewer/); JGI Genome Browser (http://genome.jgi-psf.org/); GBrowse (Stein et al. 2002); Anno-J (Lister et al. 2008).